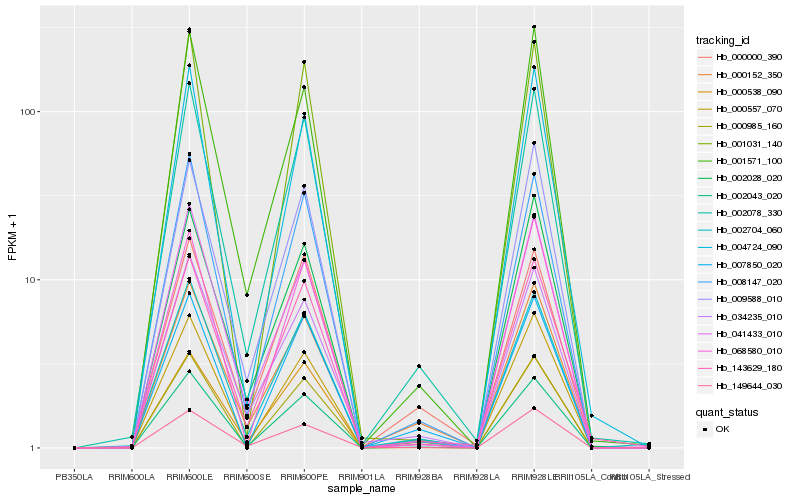
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_330 |
0.0 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 2 |
Hb_002043_020 |
0.04099468 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 3 |
Hb_002704_060 |
0.045435358 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 4 |
Hb_008147_020 |
0.0491712413 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 5 |
Hb_143629_180 |
0.0513120383 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |
| 6 |
Hb_000538_090 |
0.0606920081 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1603380 [Ricinus communis] |
| 7 |
Hb_034235_010 |
0.0667926993 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 8 |
Hb_000152_350 |
0.0679852912 |
- |
- |
PREDICTED: enolase [Jatropha curcas] |
| 9 |
Hb_149644_030 |
0.0702402754 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 10 |
Hb_000985_160 |
0.0716286308 |
- |
- |
PREDICTED: cytochrome P450 724B1-like [Jatropha curcas] |
| 11 |
Hb_000557_070 |
0.0720840397 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 12 |
Hb_002028_020 |
0.0728745808 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 13 |
Hb_041433_010 |
0.0747259554 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_004724_090 |
0.0747304153 |
- |
- |
PREDICTED: uncharacterized protein LOC105638638 [Jatropha curcas] |
| 15 |
Hb_000000_390 |
0.0768694386 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |
| 16 |
Hb_009588_010 |
0.0770261875 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 17 |
Hb_001571_100 |
0.0774503902 |
- |
- |
chlorophyll A-B binding family protein [Populus trichocarpa] |
| 18 |
Hb_007850_020 |
0.0784674742 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001031_140 |
0.0789656532 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g33370-like [Jatropha curcas] |
| 20 |
Hb_068580_010 |
0.0792857555 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 isoform X2 [Populus euphratica] |