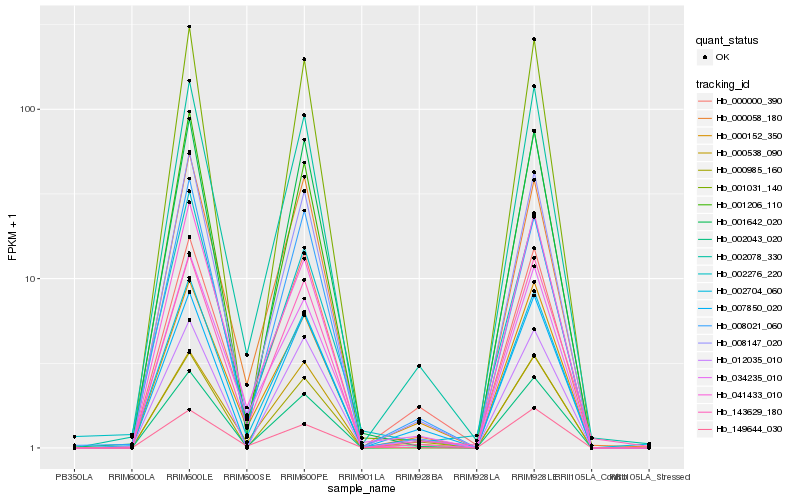
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002704_060 |
0.0 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 2 |
Hb_002078_330 |
0.045435358 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 3 |
Hb_008147_020 |
0.053254135 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 4 |
Hb_002043_020 |
0.0584788786 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 5 |
Hb_034235_010 |
0.0654623563 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 6 |
Hb_000538_090 |
0.0741027716 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1603380 [Ricinus communis] |
| 7 |
Hb_143629_180 |
0.0746436783 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |
| 8 |
Hb_000152_350 |
0.080403288 |
- |
- |
PREDICTED: enolase [Jatropha curcas] |
| 9 |
Hb_041433_010 |
0.0829347995 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_000058_180 |
0.0833923202 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |
| 11 |
Hb_000000_390 |
0.0846231183 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |
| 12 |
Hb_008021_060 |
0.0849084622 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 13 |
Hb_001206_110 |
0.0852694826 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45910 [Jatropha curcas] |
| 14 |
Hb_149644_030 |
0.0860282598 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 15 |
Hb_000985_160 |
0.086802975 |
- |
- |
PREDICTED: cytochrome P450 724B1-like [Jatropha curcas] |
| 16 |
Hb_001031_140 |
0.0883688261 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g33370-like [Jatropha curcas] |
| 17 |
Hb_012035_010 |
0.0898813115 |
- |
- |
hypothetical protein JCGZ_21143 [Jatropha curcas] |
| 18 |
Hb_002276_220 |
0.0905212505 |
- |
- |
PREDICTED: uncharacterized protein LOC105640684 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001642_020 |
0.0912983852 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 9 [Jatropha curcas] |
| 20 |
Hb_007850_020 |
0.0923827491 |
- |
- |
conserved hypothetical protein [Ricinus communis] |