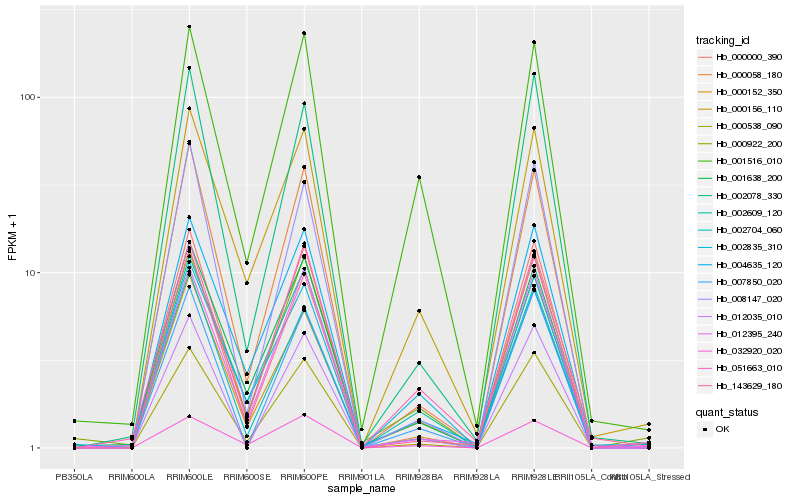
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000000_390 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |
| 2 |
Hb_000538_090 |
0.0481539841 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1603380 [Ricinus communis] |
| 3 |
Hb_051663_010 |
0.0752187459 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 4 |
Hb_004635_120 |
0.0755675421 |
- |
- |
PREDICTED: rho GTPase-activating protein 4-like [Jatropha curcas] |
| 5 |
Hb_012395_240 |
0.0767889511 |
- |
- |
PREDICTED: uncharacterized protein LOC105638421 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002078_330 |
0.0768694386 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 7 |
Hb_001638_200 |
0.0776405145 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 43 [Jatropha curcas] |
| 8 |
Hb_000156_110 |
0.0787692226 |
- |
- |
Uncharacterized protein TCM_029490 [Theobroma cacao] |
| 9 |
Hb_000922_200 |
0.0793629232 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0012s08280g [Populus trichocarpa] |
| 10 |
Hb_032920_020 |
0.0800921745 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000152_350 |
0.0801275907 |
- |
- |
PREDICTED: enolase [Jatropha curcas] |
| 12 |
Hb_002835_310 |
0.080988138 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_012035_010 |
0.0835597498 |
- |
- |
hypothetical protein JCGZ_21143 [Jatropha curcas] |
| 14 |
Hb_002704_060 |
0.0846231183 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 15 |
Hb_007850_020 |
0.086860782 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001516_010 |
0.0878842021 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 17 |
Hb_002609_120 |
0.0881754809 |
- |
- |
hypothetical protein POPTR_0001s25660g [Populus trichocarpa] |
| 18 |
Hb_000058_180 |
0.0882303086 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |
| 19 |
Hb_008147_020 |
0.0892197907 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 20 |
Hb_143629_180 |
0.0907251673 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |