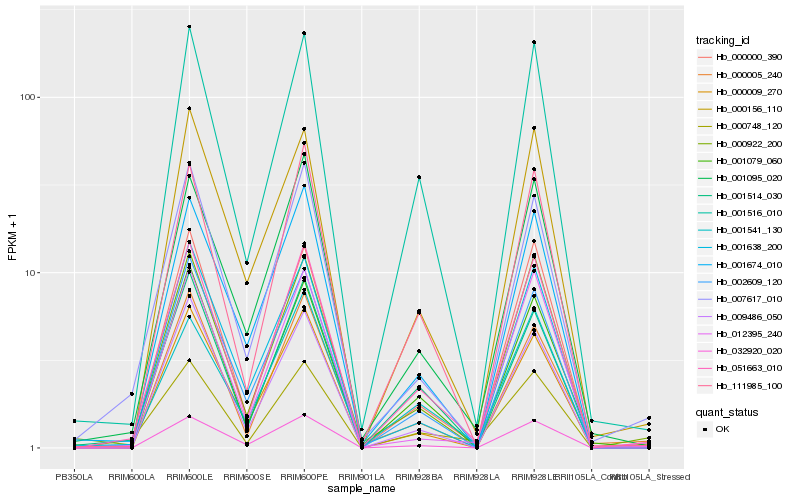
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_051663_010 |
0.0 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 2 |
Hb_001516_010 |
0.0636178533 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 3 |
Hb_000000_390 |
0.0752187459 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |
| 4 |
Hb_111985_100 |
0.0768662784 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 5 |
Hb_000922_200 |
0.0790403575 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0012s08280g [Populus trichocarpa] |
| 6 |
Hb_032920_020 |
0.0831501925 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_009486_050 |
0.0881625313 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 8 |
Hb_007617_010 |
0.0891005827 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001079_060 |
0.0912047797 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001095_020 |
0.0917268089 |
- |
- |
PREDICTED: F-box protein At2g26850-like [Jatropha curcas] |
| 11 |
Hb_000156_110 |
0.0943963805 |
- |
- |
Uncharacterized protein TCM_029490 [Theobroma cacao] |
| 12 |
Hb_002609_120 |
0.0945119298 |
- |
- |
hypothetical protein POPTR_0001s25660g [Populus trichocarpa] |
| 13 |
Hb_001514_030 |
0.0957976671 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 14 |
Hb_001541_130 |
0.0963283003 |
- |
- |
GDSL esterase/lipase [Morus notabilis] |
| 15 |
Hb_000009_270 |
0.0967438854 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 16 |
Hb_001674_010 |
0.0975191503 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 17 |
Hb_000005_240 |
0.0982590544 |
- |
- |
Shugoshin-1, putative [Ricinus communis] |
| 18 |
Hb_012395_240 |
0.0984962126 |
- |
- |
PREDICTED: uncharacterized protein LOC105638421 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000748_120 |
0.0994613369 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL1 [Jatropha curcas] |
| 20 |
Hb_001638_200 |
0.1003453636 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 43 [Jatropha curcas] |