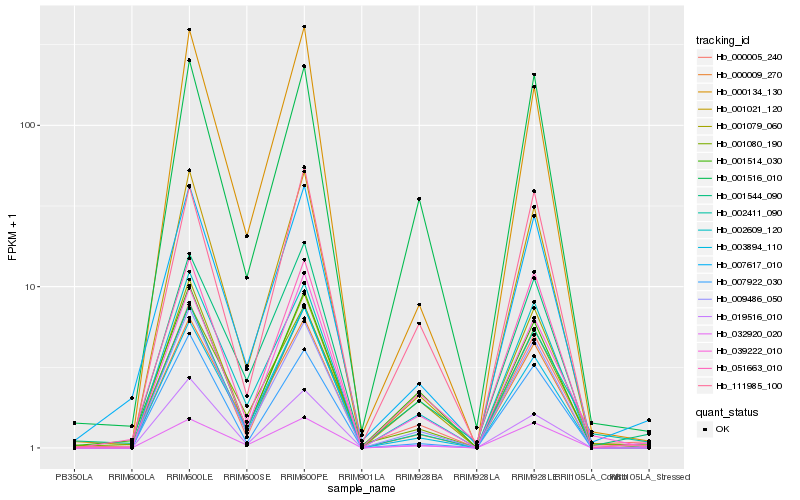
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000005_240 |
0.0 |
- |
- |
Shugoshin-1, putative [Ricinus communis] |
| 2 |
Hb_009486_050 |
0.0698762171 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 3 |
Hb_002411_090 |
0.0701487808 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL69 [Jatropha curcas] |
| 4 |
Hb_039222_010 |
0.0862630724 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001080_190 |
0.0875770416 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001021_120 |
0.0902891593 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-4 [Jatropha curcas] |
| 7 |
Hb_001079_060 |
0.0910905198 |
- |
- |
PREDICTED: probable vacuolar amino acid transporter YPQ1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007617_010 |
0.0917088507 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000009_270 |
0.093208503 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 10 |
Hb_051663_010 |
0.0982590544 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 11 |
Hb_002609_120 |
0.1007310454 |
- |
- |
hypothetical protein POPTR_0001s25660g [Populus trichocarpa] |
| 12 |
Hb_007922_030 |
0.1025536599 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |
| 13 |
Hb_003894_110 |
0.1068310155 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 14 |
Hb_000134_130 |
0.1069852645 |
- |
- |
PREDICTED: uncharacterized protein LOC105628670 [Jatropha curcas] |
| 15 |
Hb_001544_090 |
0.1075222988 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 16 |
Hb_111985_100 |
0.1083593864 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 17 |
Hb_001516_010 |
0.1102302746 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 18 |
Hb_001514_030 |
0.1106887551 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 19 |
Hb_032920_020 |
0.111600513 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 20 |
Hb_019516_010 |
0.1119213898 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like isoform X2 [Populus euphratica] |