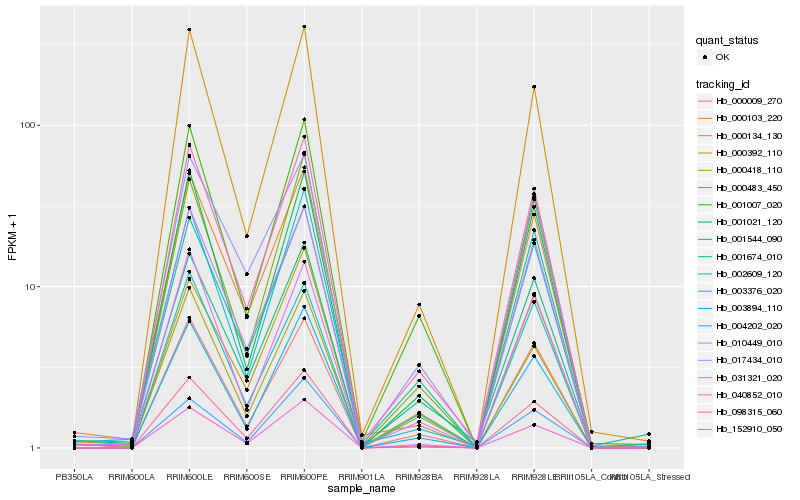
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_098315_060 |
0.0 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 2 |
Hb_152910_050 |
0.0315420953 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_040852_010 |
0.0427967386 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000134_130 |
0.0481758634 |
- |
- |
PREDICTED: uncharacterized protein LOC105628670 [Jatropha curcas] |
| 5 |
Hb_003894_110 |
0.0489647093 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 6 |
Hb_017434_010 |
0.055180622 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 7 |
Hb_000103_220 |
0.0599179913 |
- |
- |
PREDICTED: uncharacterized protein LOC105632036 [Jatropha curcas] |
| 8 |
Hb_000009_270 |
0.0602489098 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 9 |
Hb_001021_120 |
0.0636879061 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-4 [Jatropha curcas] |
| 10 |
Hb_003376_020 |
0.0639855729 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_010449_010 |
0.0741531614 |
- |
- |
l-ascorbate oxidase, putative [Ricinus communis] |
| 12 |
Hb_000483_450 |
0.0781172309 |
- |
- |
PREDICTED: anthranilate N-benzoyltransferase protein 1 [Jatropha curcas] |
| 13 |
Hb_004202_020 |
0.0801901785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000418_110 |
0.0820644505 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |
| 15 |
Hb_001007_020 |
0.0844230326 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000392_110 |
0.0864025863 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 17 |
Hb_001544_090 |
0.0878847286 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 18 |
Hb_031321_020 |
0.0882891498 |
- |
- |
hypothetical protein POPTR_0003s15520g [Populus trichocarpa] |
| 19 |
Hb_002609_120 |
0.088719028 |
- |
- |
hypothetical protein POPTR_0001s25660g [Populus trichocarpa] |
| 20 |
Hb_001674_010 |
0.0896308482 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |