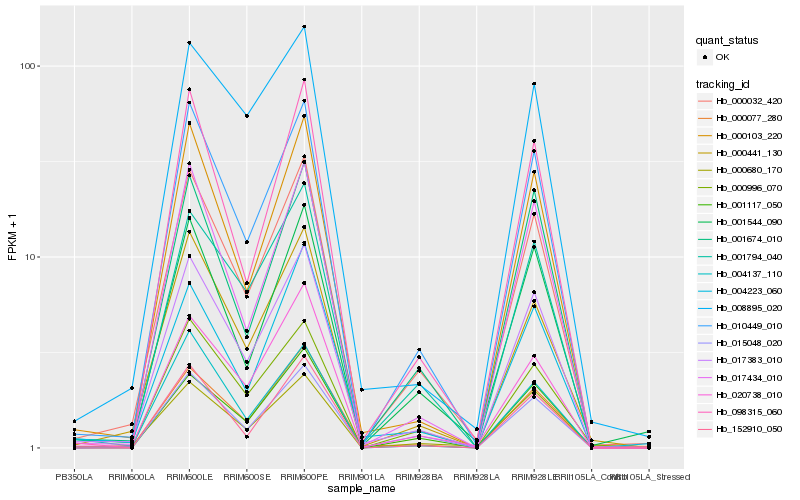
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017383_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628176 [Jatropha curcas] |
| 2 |
Hb_010449_010 |
0.0509491253 |
- |
- |
l-ascorbate oxidase, putative [Ricinus communis] |
| 3 |
Hb_000032_420 |
0.0559078478 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 4 |
Hb_000103_220 |
0.0667880624 |
- |
- |
PREDICTED: uncharacterized protein LOC105632036 [Jatropha curcas] |
| 5 |
Hb_000077_280 |
0.0679719496 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_017434_010 |
0.0782825651 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 7 |
Hb_001794_040 |
0.0792578869 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000441_130 |
0.0824539144 |
- |
- |
PREDICTED: homeobox-leucine zipper protein HOX32 [Vitis vinifera] |
| 9 |
Hb_000680_170 |
0.0879244414 |
- |
- |
hypothetical protein JCGZ_24789 [Jatropha curcas] |
| 10 |
Hb_000996_070 |
0.0915411423 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_020738_010 |
0.0916474125 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 12 |
Hb_098315_060 |
0.0926165053 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 13 |
Hb_001674_010 |
0.0957583185 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 14 |
Hb_004223_060 |
0.0958802116 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 15 |
Hb_015048_020 |
0.0982409831 |
- |
- |
PREDICTED: myb-like protein D [Jatropha curcas] |
| 16 |
Hb_008895_020 |
0.0991401678 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 17 |
Hb_001544_090 |
0.0995942695 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 18 |
Hb_152910_050 |
0.0999382759 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001117_050 |
0.100158235 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 20 |
Hb_004137_110 |
0.1003313694 |
- |
- |
PREDICTED: probable auxin efflux carrier component 6 [Jatropha curcas] |