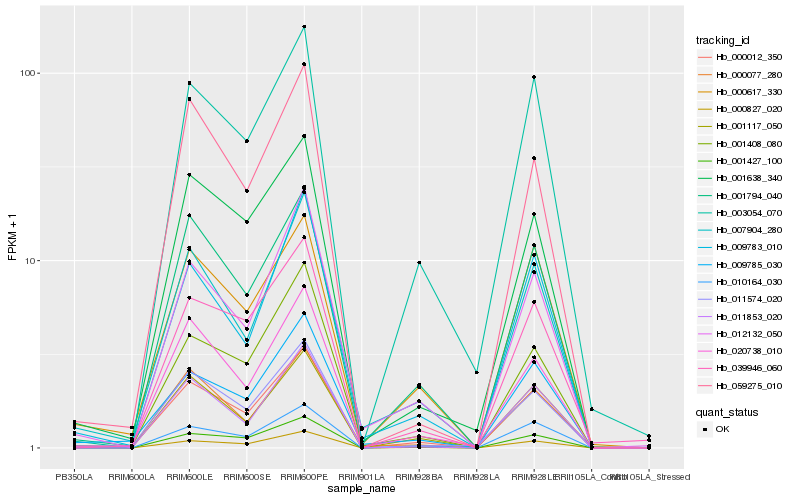
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000012_350 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein ATH1 [Jatropha curcas] |
| 2 |
Hb_001794_040 |
0.0691038033 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 3 |
Hb_011574_020 |
0.0752262981 |
- |
- |
hypothetical protein JCGZ_21524 [Jatropha curcas] |
| 4 |
Hb_010164_030 |
0.0791654308 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_020738_010 |
0.0807887812 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 6 |
Hb_001427_100 |
0.0809373516 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 7 |
Hb_000827_020 |
0.0813757033 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_007904_280 |
0.0828052732 |
- |
- |
PREDICTED: uncharacterized protein LOC105644987 [Jatropha curcas] |
| 9 |
Hb_001117_050 |
0.0851270372 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 10 |
Hb_003054_070 |
0.0880521776 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_001408_080 |
0.0901794017 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 12 |
Hb_001638_340 |
0.0904955742 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_009785_030 |
0.09161877 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |
| 14 |
Hb_000617_330 |
0.0931883437 |
- |
- |
PREDICTED: uncharacterized protein LOC105647507 [Jatropha curcas] |
| 15 |
Hb_009783_010 |
0.0934998108 |
- |
- |
PREDICTED: sulfated surface glycoprotein 185-like [Populus euphratica] |
| 16 |
Hb_012132_050 |
0.0943822013 |
- |
- |
PREDICTED: aspartic proteinase PCS1 [Jatropha curcas] |
| 17 |
Hb_039946_060 |
0.0967164888 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 18 |
Hb_011853_020 |
0.099021335 |
- |
- |
Gamma-glutamyl-gamma-aminobutyrate hydrolase, putative [Ricinus communis] |
| 19 |
Hb_000077_280 |
0.0999811467 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_059275_010 |
0.1019071465 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |