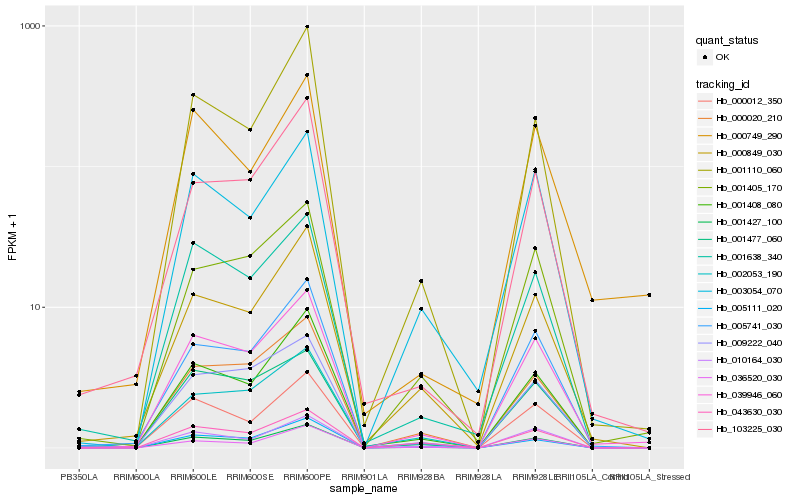
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_039946_060 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 2 |
Hb_001427_100 |
0.0627107931 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 3 |
Hb_005741_030 |
0.0889783809 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 4 |
Hb_001405_170 |
0.0927073378 |
- |
- |
aspartyl protease family protein [Populus trichocarpa] |
| 5 |
Hb_000020_210 |
0.0948550085 |
- |
- |
hypothetical protein JCGZ_17348 [Jatropha curcas] |
| 6 |
Hb_000012_350 |
0.0967164888 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein ATH1 [Jatropha curcas] |
| 7 |
Hb_003054_070 |
0.0986256122 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_001408_080 |
0.0989275677 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 9 |
Hb_010164_030 |
0.1009524785 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000849_030 |
0.1028534754 |
- |
- |
PREDICTED: fimbrin-2 [Jatropha curcas] |
| 11 |
Hb_009222_040 |
0.1035425619 |
transcription factor |
TF Family: C2C2-Dof |
DNA binding with one finger 2.4, putative [Theobroma cacao] |
| 12 |
Hb_001638_340 |
0.1040144349 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005111_020 |
0.1056453195 |
- |
- |
hypothetical protein CISIN_1g0417821mg, partial [Citrus sinensis] |
| 14 |
Hb_001477_060 |
0.1064791092 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 15 |
Hb_002053_190 |
0.1094013153 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 16 |
Hb_036520_030 |
0.1106658205 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |
| 17 |
Hb_000749_290 |
0.1124586517 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 18 |
Hb_043630_030 |
0.1136439402 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 19 |
Hb_001110_060 |
0.1143336819 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 20 |
Hb_103225_030 |
0.1146459713 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |