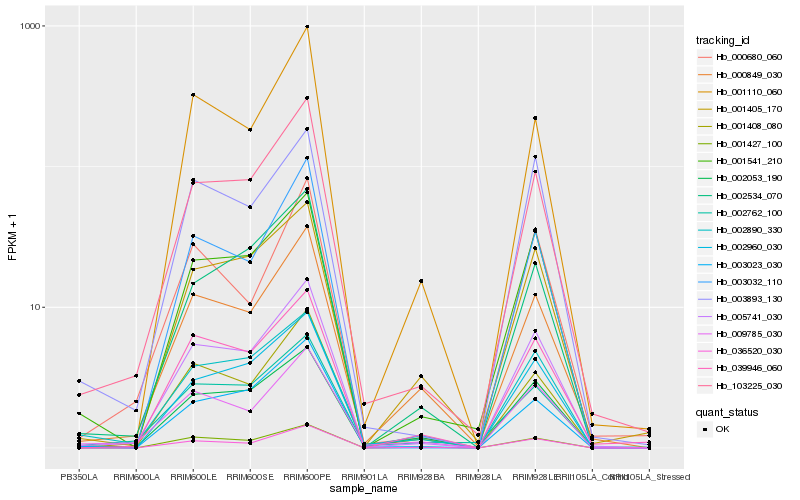
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005741_030 |
0.0 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 2 |
Hb_103225_030 |
0.0588784194 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_001408_080 |
0.0836795435 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 4 |
Hb_001427_100 |
0.0878521316 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 5 |
Hb_036520_030 |
0.0882766069 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |
| 6 |
Hb_001541_210 |
0.0886607115 |
- |
- |
hypothetical protein POPTR_0008s19020g [Populus trichocarpa] |
| 7 |
Hb_039946_060 |
0.0889783809 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 8 |
Hb_003032_110 |
0.0892130368 |
- |
- |
- |
| 9 |
Hb_002960_030 |
0.0955120802 |
- |
- |
hypothetical protein POPTR_0002s02190g [Populus trichocarpa] |
| 10 |
Hb_003893_130 |
0.0965376464 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 11 |
Hb_000849_030 |
0.0968941905 |
- |
- |
PREDICTED: fimbrin-2 [Jatropha curcas] |
| 12 |
Hb_002053_190 |
0.100800995 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002890_330 |
0.102154987 |
- |
- |
caspase, putative [Ricinus communis] |
| 14 |
Hb_003023_030 |
0.105438524 |
- |
- |
PREDICTED: MATE efflux family protein 5-like isoform X1 [Citrus sinensis] |
| 15 |
Hb_009785_030 |
0.1059566187 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |
| 16 |
Hb_001405_170 |
0.1084598438 |
- |
- |
aspartyl protease family protein [Populus trichocarpa] |
| 17 |
Hb_002762_100 |
0.1091874774 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_002534_070 |
0.1094835708 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 19 |
Hb_000680_060 |
0.1114014494 |
- |
- |
hypothetical protein POPTR_0001s20790g [Populus trichocarpa] |
| 20 |
Hb_001110_060 |
0.1123349405 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |