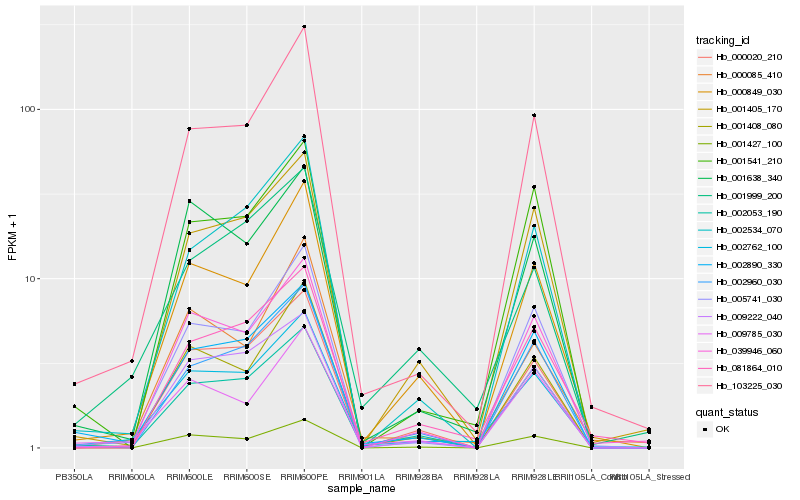
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002762_100 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_103225_030 |
0.0907424409 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_005741_030 |
0.1091874774 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 4 |
Hb_000849_030 |
0.1095077435 |
- |
- |
PREDICTED: fimbrin-2 [Jatropha curcas] |
| 5 |
Hb_002890_330 |
0.1100604403 |
- |
- |
caspase, putative [Ricinus communis] |
| 6 |
Hb_081864_010 |
0.1151036886 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000020_210 |
0.1159996083 |
- |
- |
hypothetical protein JCGZ_17348 [Jatropha curcas] |
| 8 |
Hb_002053_190 |
0.1167840268 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001408_080 |
0.1174508284 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 10 |
Hb_039946_060 |
0.1195426101 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 11 |
Hb_001405_170 |
0.1200459754 |
- |
- |
aspartyl protease family protein [Populus trichocarpa] |
| 12 |
Hb_009222_040 |
0.120518378 |
transcription factor |
TF Family: C2C2-Dof |
DNA binding with one finger 2.4, putative [Theobroma cacao] |
| 13 |
Hb_001999_200 |
0.1221255374 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_009785_030 |
0.1251717532 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |
| 15 |
Hb_000085_410 |
0.1255671929 |
- |
- |
hypothetical protein POPTR_0011s09040g [Populus trichocarpa] |
| 16 |
Hb_002960_030 |
0.126692549 |
- |
- |
hypothetical protein POPTR_0002s02190g [Populus trichocarpa] |
| 17 |
Hb_001638_340 |
0.1268982138 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001541_210 |
0.1285414304 |
- |
- |
hypothetical protein POPTR_0008s19020g [Populus trichocarpa] |
| 19 |
Hb_002534_070 |
0.1290147675 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 20 |
Hb_001427_100 |
0.1295427902 |
- |
- |
acc synthase, putative [Ricinus communis] |