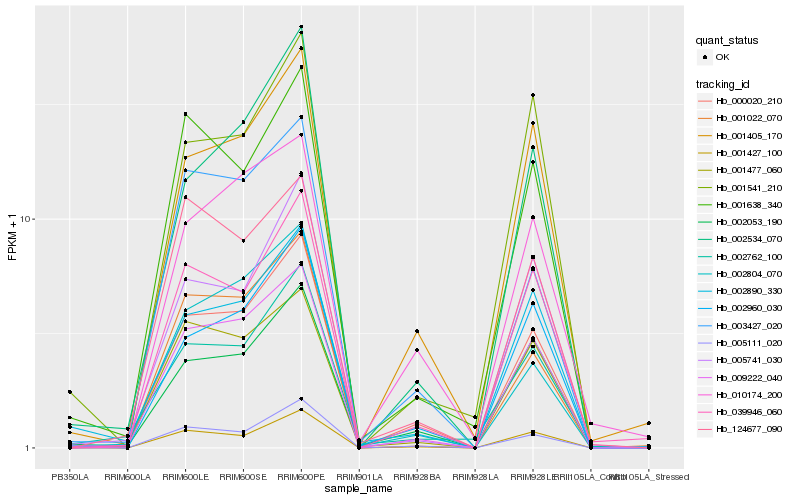
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009222_040 |
0.0 |
transcription factor |
TF Family: C2C2-Dof |
DNA binding with one finger 2.4, putative [Theobroma cacao] |
| 2 |
Hb_000020_210 |
0.0661497639 |
- |
- |
hypothetical protein JCGZ_17348 [Jatropha curcas] |
| 3 |
Hb_001477_060 |
0.0802919617 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 4 |
Hb_001405_170 |
0.0952253964 |
- |
- |
aspartyl protease family protein [Populus trichocarpa] |
| 5 |
Hb_002053_190 |
0.0992087117 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002890_330 |
0.0994187823 |
- |
- |
caspase, putative [Ricinus communis] |
| 7 |
Hb_001427_100 |
0.1018550115 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 8 |
Hb_039946_060 |
0.1035425619 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 9 |
Hb_002960_030 |
0.1039351101 |
- |
- |
hypothetical protein POPTR_0002s02190g [Populus trichocarpa] |
| 10 |
Hb_001022_070 |
0.1066627096 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 11 |
Hb_001638_340 |
0.1088030391 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002534_070 |
0.1098235561 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 13 |
Hb_001541_210 |
0.1156307178 |
- |
- |
hypothetical protein POPTR_0008s19020g [Populus trichocarpa] |
| 14 |
Hb_124677_090 |
0.1163754091 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005741_030 |
0.1178003709 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 16 |
Hb_005111_020 |
0.1182976245 |
- |
- |
hypothetical protein CISIN_1g0417821mg, partial [Citrus sinensis] |
| 17 |
Hb_003427_020 |
0.1191020131 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002762_100 |
0.120518378 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_010174_200 |
0.1212162168 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002804_070 |
0.1221194549 |
- |
- |
hypothetical protein JCGZ_05701 [Jatropha curcas] |