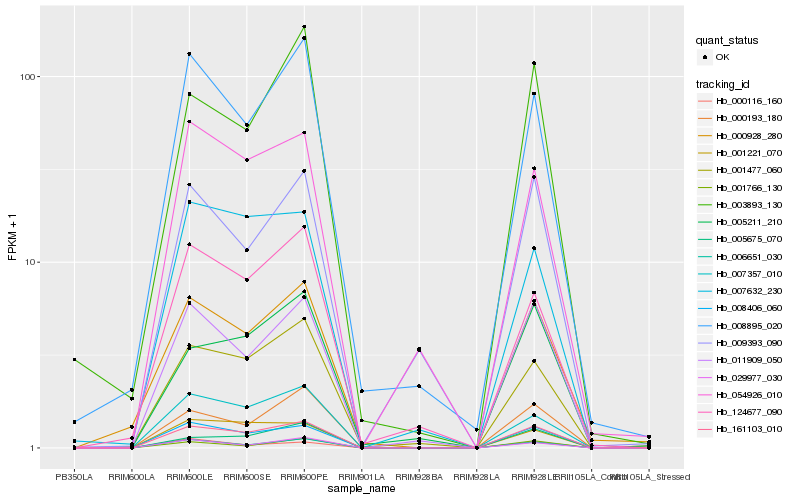
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_161103_010 |
0.0 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 2 |
Hb_008406_060 |
0.0619753516 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 1-like [Jatropha curcas] |
| 3 |
Hb_011909_050 |
0.0900756361 |
- |
- |
PREDICTED: calcium-binding protein PBP1 [Jatropha curcas] |
| 4 |
Hb_006651_030 |
0.0959798136 |
- |
- |
unnamed protein product [Homo sapiens] |
| 5 |
Hb_001477_060 |
0.1006202003 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 6 |
Hb_007357_010 |
0.1074293283 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_000928_280 |
0.1168368796 |
- |
- |
hypothetical protein VITISV_005300 [Vitis vinifera] |
| 8 |
Hb_000193_180 |
0.1177889697 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB28-like [Jatropha curcas] |
| 9 |
Hb_005675_070 |
0.1183978663 |
- |
- |
hypothetical protein EUGRSUZ_K01585 [Eucalyptus grandis] |
| 10 |
Hb_008895_020 |
0.1213137934 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 11 |
Hb_001221_070 |
0.1264339425 |
- |
- |
PREDICTED: calcium-binding protein CML38-like [Jatropha curcas] |
| 12 |
Hb_001766_130 |
0.127038113 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_029977_030 |
0.1277081323 |
transcription factor |
TF Family: bZIP |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_054926_010 |
0.1322445159 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 15 |
Hb_003893_130 |
0.1329616263 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 16 |
Hb_000116_160 |
0.1336152322 |
transcription factor |
TF Family: RWP-RK |
hypothetical protein JCGZ_22391 [Jatropha curcas] |
| 17 |
Hb_005211_210 |
0.135329691 |
- |
- |
lysine and histidine specific transporter family protein [Populus trichocarpa] |
| 18 |
Hb_007632_230 |
0.1367955457 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-3-like [Jatropha curcas] |
| 19 |
Hb_009393_090 |
0.1369937974 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 20 |
Hb_124677_090 |
0.1370614568 |
- |
- |
conserved hypothetical protein [Ricinus communis] |