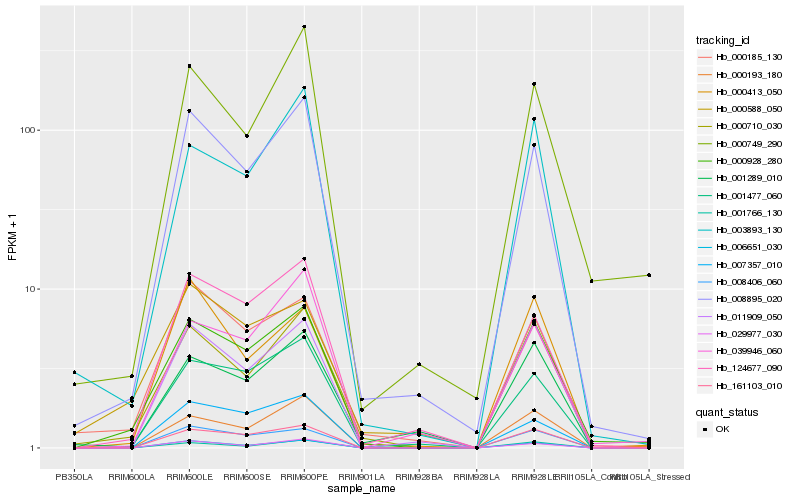
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000928_280 |
0.0 |
- |
- |
hypothetical protein VITISV_005300 [Vitis vinifera] |
| 2 |
Hb_000710_030 |
0.1107062438 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 3 |
Hb_161103_010 |
0.1168368796 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 4 |
Hb_008895_020 |
0.1179484886 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 5 |
Hb_008406_060 |
0.1302249827 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 1-like [Jatropha curcas] |
| 6 |
Hb_006651_030 |
0.1346001728 |
- |
- |
unnamed protein product [Homo sapiens] |
| 7 |
Hb_001477_060 |
0.1351514154 |
transcription factor |
TF Family: C2H2 |
zinc finger family protein [Populus trichocarpa] |
| 8 |
Hb_000193_180 |
0.1361926631 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB28-like [Jatropha curcas] |
| 9 |
Hb_011909_050 |
0.1369732333 |
- |
- |
PREDICTED: calcium-binding protein PBP1 [Jatropha curcas] |
| 10 |
Hb_000185_130 |
0.1421312375 |
- |
- |
UBX domain-containing protein 8-B, putative [Ricinus communis] |
| 11 |
Hb_124677_090 |
0.1450259486 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003893_130 |
0.1452211881 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 13 |
Hb_000749_290 |
0.1519384746 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 14 |
Hb_029977_030 |
0.1541350558 |
transcription factor |
TF Family: bZIP |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001766_130 |
0.1550364885 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_000588_050 |
0.1560378027 |
transcription factor |
TF Family: Trihelix |
hypothetical protein POPTR_0008s02580g [Populus trichocarpa] |
| 17 |
Hb_001289_010 |
0.1561062334 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Vitis vinifera] |
| 18 |
Hb_007357_010 |
0.1573161126 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_039946_060 |
0.1590881043 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 20 |
Hb_000413_050 |
0.1602643648 |
- |
- |
PREDICTED: transcription factor bHLH112 isoform X2 [Jatropha curcas] |