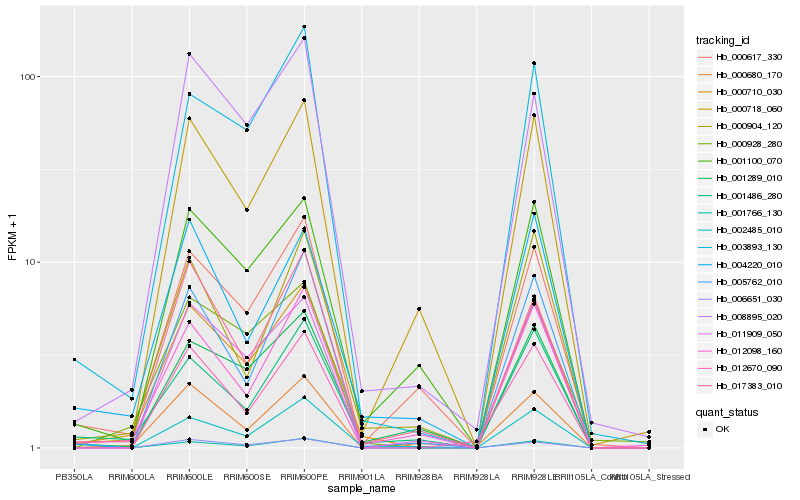
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000710_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 2 |
Hb_001766_130 |
0.1073698454 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000928_280 |
0.1107062438 |
- |
- |
hypothetical protein VITISV_005300 [Vitis vinifera] |
| 4 |
Hb_003893_130 |
0.1122214061 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 5 |
Hb_012098_160 |
0.1153548411 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 6 |
Hb_011909_050 |
0.1221557914 |
- |
- |
PREDICTED: calcium-binding protein PBP1 [Jatropha curcas] |
| 7 |
Hb_008895_020 |
0.123636391 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 8 |
Hb_001486_280 |
0.1238235361 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |
| 9 |
Hb_006651_030 |
0.124042839 |
- |
- |
unnamed protein product [Homo sapiens] |
| 10 |
Hb_001289_010 |
0.1265872446 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Vitis vinifera] |
| 11 |
Hb_005762_010 |
0.129494169 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic-like [Populus euphratica] |
| 12 |
Hb_017383_010 |
0.1316186907 |
- |
- |
PREDICTED: uncharacterized protein LOC105628176 [Jatropha curcas] |
| 13 |
Hb_000904_120 |
0.1316843057 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001100_070 |
0.1329664739 |
- |
- |
PREDICTED: ferredoxin--nitrite reductase, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_004220_010 |
0.1337027828 |
- |
- |
PREDICTED: uncharacterized protein LOC105642366 [Jatropha curcas] |
| 16 |
Hb_000617_330 |
0.1339562491 |
- |
- |
PREDICTED: uncharacterized protein LOC105647507 [Jatropha curcas] |
| 17 |
Hb_002485_010 |
0.1340351339 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 18 |
Hb_012670_090 |
0.135041758 |
- |
- |
GDSL-motif lipase/hydrolase family protein [Populus trichocarpa] |
| 19 |
Hb_000680_170 |
0.1354578465 |
- |
- |
hypothetical protein JCGZ_24789 [Jatropha curcas] |
| 20 |
Hb_000718_060 |
0.1356027142 |
transcription factor |
TF Family: MYB |
DNA binding protein, putative [Ricinus communis] |