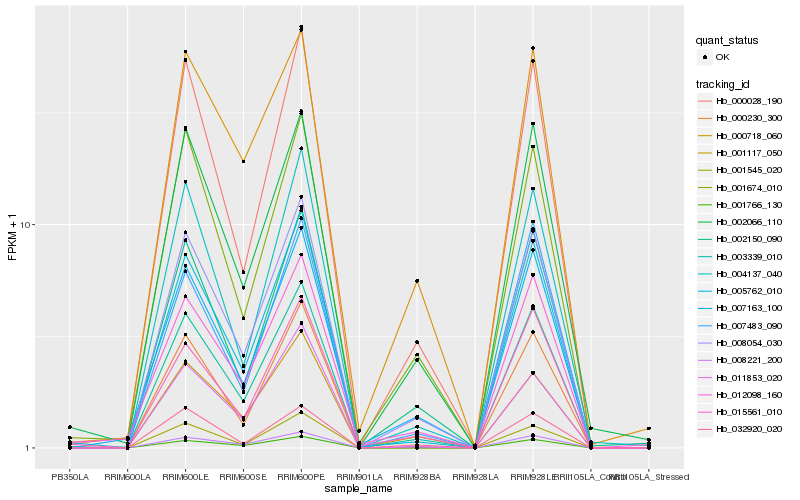
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003339_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_04276 [Jatropha curcas] |
| 2 |
Hb_008054_030 |
0.0188899738 |
- |
- |
PREDICTED: VQ motif-containing protein 1-like [Jatropha curcas] |
| 3 |
Hb_005762_010 |
0.0542241431 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic-like [Populus euphratica] |
| 4 |
Hb_000028_190 |
0.0646258282 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 5 |
Hb_007163_100 |
0.0690874822 |
- |
- |
BnaC03g52280D [Brassica napus] |
| 6 |
Hb_002150_090 |
0.0741410578 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 7 |
Hb_004137_040 |
0.0747193799 |
- |
- |
PREDICTED: serine carboxypeptidase-like 42 [Jatropha curcas] |
| 8 |
Hb_001766_130 |
0.0767504119 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_008221_200 |
0.078129781 |
- |
- |
PREDICTED: protein trichome birefringence-like 41 [Jatropha curcas] |
| 10 |
Hb_002066_110 |
0.0795702763 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 11 |
Hb_000230_300 |
0.0801743259 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme 4 [Jatropha curcas] |
| 12 |
Hb_001674_010 |
0.0857893869 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 13 |
Hb_001117_050 |
0.0865082116 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 14 |
Hb_000718_060 |
0.0874528554 |
transcription factor |
TF Family: MYB |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_012098_160 |
0.0899078536 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 16 |
Hb_011853_020 |
0.091945897 |
- |
- |
Gamma-glutamyl-gamma-aminobutyrate hydrolase, putative [Ricinus communis] |
| 17 |
Hb_007483_090 |
0.0919830116 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001545_020 |
0.0932508215 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X5 [Populus euphratica] |
| 19 |
Hb_015561_010 |
0.0936155374 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_032920_020 |
0.0944707735 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |