| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
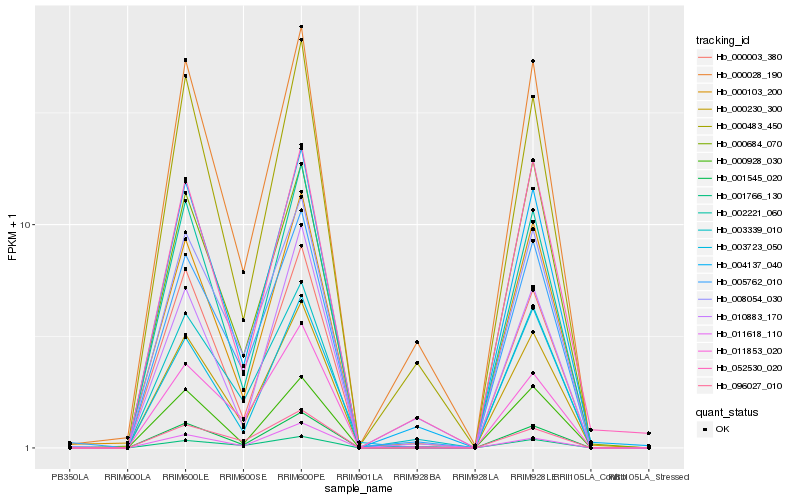
Hb_000230_300 |
0.0 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme 4 [Jatropha curcas] |
| 2 |
Hb_002221_060 |
0.0446201431 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 3 |
Hb_000003_380 |
0.0564576824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004137_040 |
0.0632070543 |
- |
- |
PREDICTED: serine carboxypeptidase-like 42 [Jatropha curcas] |
| 5 |
Hb_005762_010 |
0.063385269 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic-like [Populus euphratica] |
| 6 |
Hb_000103_200 |
0.065765513 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 7 |
Hb_000028_190 |
0.0794993827 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 8 |
Hb_000928_030 |
0.0798954997 |
transcription factor |
TF Family: bZIP |
PREDICTED: protein ABSCISIC ACID-INSENSITIVE 5 [Jatropha curcas] |
| 9 |
Hb_003339_010 |
0.0801743259 |
- |
- |
hypothetical protein JCGZ_04276 [Jatropha curcas] |
| 10 |
Hb_000684_070 |
0.0862980586 |
- |
- |
PREDICTED: uncharacterized protein LOC105642198 isoform X1 [Jatropha curcas] |
| 11 |
Hb_008054_030 |
0.0878416736 |
- |
- |
PREDICTED: VQ motif-containing protein 1-like [Jatropha curcas] |
| 12 |
Hb_000483_450 |
0.0883935857 |
- |
- |
PREDICTED: anthranilate N-benzoyltransferase protein 1 [Jatropha curcas] |
| 13 |
Hb_001766_130 |
0.0886765787 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_096027_010 |
0.0910931229 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_003723_050 |
0.0922149026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_010883_170 |
0.0923513364 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 17 |
Hb_011853_020 |
0.0930649822 |
- |
- |
Gamma-glutamyl-gamma-aminobutyrate hydrolase, putative [Ricinus communis] |
| 18 |
Hb_001545_020 |
0.0934025749 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X5 [Populus euphratica] |
| 19 |
Hb_011618_110 |
0.0984886766 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020-like [Citrus sinensis] |
| 20 |
Hb_052530_020 |
0.0985197501 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |