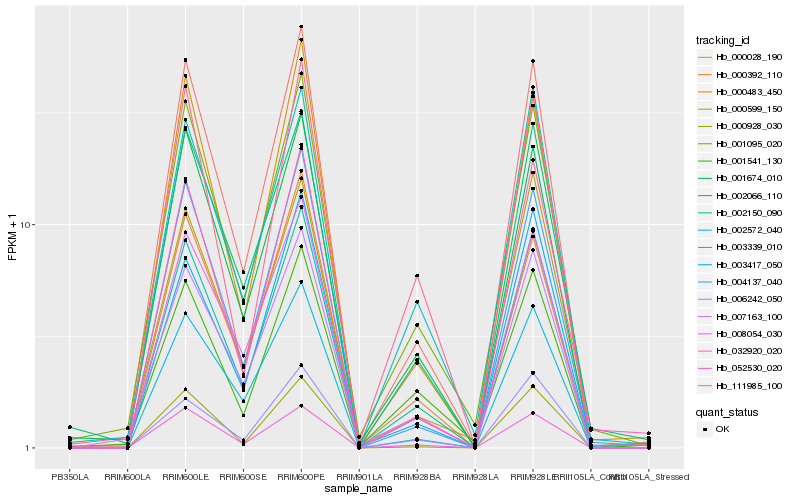
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002150_090 |
0.0 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 2 |
Hb_000028_190 |
0.0536723714 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 3 |
Hb_001095_020 |
0.0584332191 |
- |
- |
PREDICTED: F-box protein At2g26850-like [Jatropha curcas] |
| 4 |
Hb_007163_100 |
0.0622286132 |
- |
- |
BnaC03g52280D [Brassica napus] |
| 5 |
Hb_008054_030 |
0.0665986579 |
- |
- |
PREDICTED: VQ motif-containing protein 1-like [Jatropha curcas] |
| 6 |
Hb_002066_110 |
0.0670055433 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 7 |
Hb_001674_010 |
0.0681912996 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 8 |
Hb_052530_020 |
0.0686690794 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 9 |
Hb_002572_040 |
0.0710464376 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 10 |
Hb_032920_020 |
0.0713051325 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_004137_040 |
0.0723290706 |
- |
- |
PREDICTED: serine carboxypeptidase-like 42 [Jatropha curcas] |
| 12 |
Hb_003339_010 |
0.0741410578 |
- |
- |
hypothetical protein JCGZ_04276 [Jatropha curcas] |
| 13 |
Hb_000599_150 |
0.0813598214 |
- |
- |
PREDICTED: probable arabinose 5-phosphate isomerase [Jatropha curcas] |
| 14 |
Hb_000392_110 |
0.0844348829 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 15 |
Hb_001541_130 |
0.0850394536 |
- |
- |
GDSL esterase/lipase [Morus notabilis] |
| 16 |
Hb_003417_050 |
0.0852149706 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_006242_050 |
0.0856426157 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |
| 18 |
Hb_000483_450 |
0.0875859494 |
- |
- |
PREDICTED: anthranilate N-benzoyltransferase protein 1 [Jatropha curcas] |
| 19 |
Hb_000928_030 |
0.088975284 |
transcription factor |
TF Family: bZIP |
PREDICTED: protein ABSCISIC ACID-INSENSITIVE 5 [Jatropha curcas] |
| 20 |
Hb_111985_100 |
0.0923675438 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |