| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
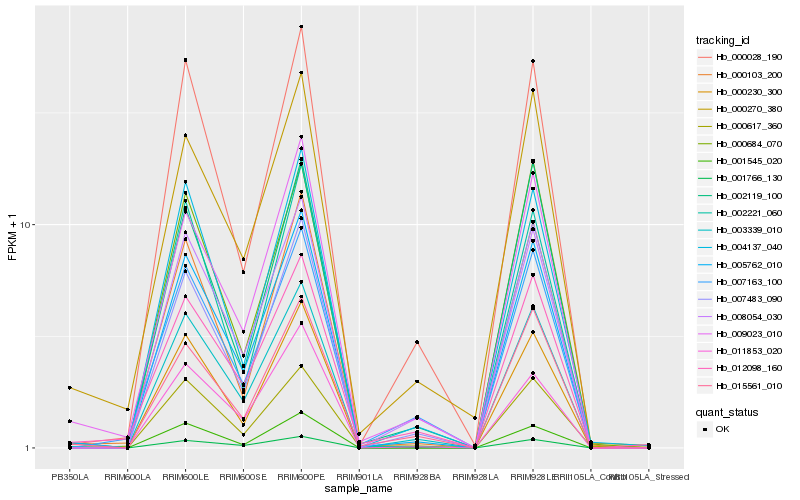
Hb_005762_010 |
0.0 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic-like [Populus euphratica] |
| 2 |
Hb_003339_010 |
0.0542241431 |
- |
- |
hypothetical protein JCGZ_04276 [Jatropha curcas] |
| 3 |
Hb_000230_300 |
0.063385269 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme 4 [Jatropha curcas] |
| 4 |
Hb_008054_030 |
0.0652682097 |
- |
- |
PREDICTED: VQ motif-containing protein 1-like [Jatropha curcas] |
| 5 |
Hb_000103_200 |
0.0701015742 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 6 |
Hb_009023_010 |
0.0725862234 |
- |
- |
glycosyl hydrolase family 31 family protein [Populus trichocarpa] |
| 7 |
Hb_004137_040 |
0.0746513897 |
- |
- |
PREDICTED: serine carboxypeptidase-like 42 [Jatropha curcas] |
| 8 |
Hb_001766_130 |
0.0760361604 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_001545_020 |
0.0768962153 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X5 [Populus euphratica] |
| 10 |
Hb_000028_190 |
0.076999354 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 11 |
Hb_015561_010 |
0.0830484477 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002119_100 |
0.084811995 |
- |
- |
cytochrome B561, putative [Ricinus communis] |
| 13 |
Hb_012098_160 |
0.0873277389 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 14 |
Hb_002221_060 |
0.0877471516 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 15 |
Hb_007163_100 |
0.0905981986 |
- |
- |
BnaC03g52280D [Brassica napus] |
| 16 |
Hb_011853_020 |
0.0908571235 |
- |
- |
Gamma-glutamyl-gamma-aminobutyrate hydrolase, putative [Ricinus communis] |
| 17 |
Hb_007483_090 |
0.0910877032 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000617_360 |
0.0927651273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000270_380 |
0.0936830884 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 20 |
Hb_000684_070 |
0.0947737918 |
- |
- |
PREDICTED: uncharacterized protein LOC105642198 isoform X1 [Jatropha curcas] |