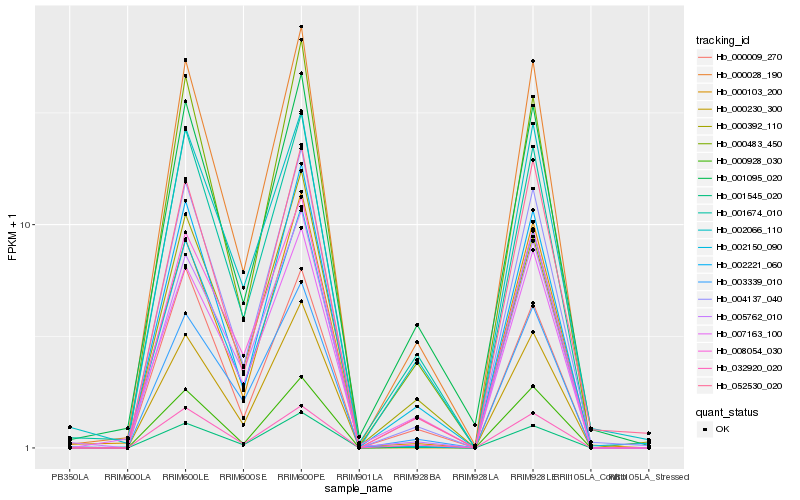
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000028_190 |
0.0 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 2 |
Hb_004137_040 |
0.0469854656 |
- |
- |
PREDICTED: serine carboxypeptidase-like 42 [Jatropha curcas] |
| 3 |
Hb_002150_090 |
0.0536723714 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 4 |
Hb_000483_450 |
0.0542545312 |
- |
- |
PREDICTED: anthranilate N-benzoyltransferase protein 1 [Jatropha curcas] |
| 5 |
Hb_001674_010 |
0.056744331 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 6 |
Hb_008054_030 |
0.0593961423 |
- |
- |
PREDICTED: VQ motif-containing protein 1-like [Jatropha curcas] |
| 7 |
Hb_000928_030 |
0.0612894758 |
transcription factor |
TF Family: bZIP |
PREDICTED: protein ABSCISIC ACID-INSENSITIVE 5 [Jatropha curcas] |
| 8 |
Hb_001095_020 |
0.0626279479 |
- |
- |
PREDICTED: F-box protein At2g26850-like [Jatropha curcas] |
| 9 |
Hb_032920_020 |
0.0638082908 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 10 |
Hb_003339_010 |
0.0646258282 |
- |
- |
hypothetical protein JCGZ_04276 [Jatropha curcas] |
| 11 |
Hb_007163_100 |
0.0677882623 |
- |
- |
BnaC03g52280D [Brassica napus] |
| 12 |
Hb_000103_200 |
0.0737161098 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 13 |
Hb_001545_020 |
0.0766806595 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X5 [Populus euphratica] |
| 14 |
Hb_005762_010 |
0.076999354 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic-like [Populus euphratica] |
| 15 |
Hb_002221_060 |
0.0771126144 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 16 |
Hb_052530_020 |
0.0778900493 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 17 |
Hb_000230_300 |
0.0794993827 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme 4 [Jatropha curcas] |
| 18 |
Hb_000392_110 |
0.0815062242 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 19 |
Hb_000009_270 |
0.0830794484 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 20 |
Hb_002066_110 |
0.0833635853 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |