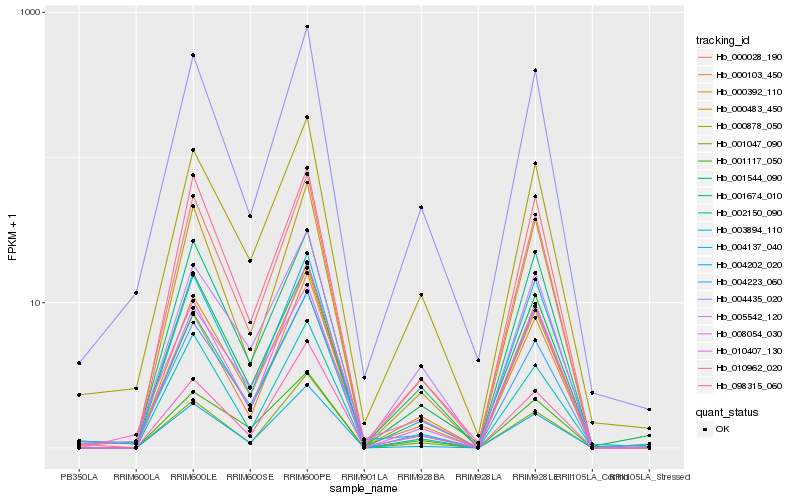
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000392_110 |
0.0 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 2 |
Hb_004202_020 |
0.0659902638 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000483_450 |
0.0698681135 |
- |
- |
PREDICTED: anthranilate N-benzoyltransferase protein 1 [Jatropha curcas] |
| 4 |
Hb_000878_050 |
0.0725331937 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 5 |
Hb_001117_050 |
0.0727273546 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 6 |
Hb_003894_110 |
0.0774996251 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 7 |
Hb_001047_090 |
0.0804099499 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP16-like [Jatropha curcas] |
| 8 |
Hb_008054_030 |
0.0812133948 |
- |
- |
PREDICTED: VQ motif-containing protein 1-like [Jatropha curcas] |
| 9 |
Hb_000028_190 |
0.0815062242 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 10 |
Hb_005542_120 |
0.0815553709 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At3g28040 [Jatropha curcas] |
| 11 |
Hb_001544_090 |
0.0830839168 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 12 |
Hb_000103_450 |
0.0832921072 |
- |
- |
PREDICTED: dentin sialophosphoprotein-like [Jatropha curcas] |
| 13 |
Hb_004435_020 |
0.0840590512 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 14 |
Hb_002150_090 |
0.0844348829 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 15 |
Hb_004137_040 |
0.0848355963 |
- |
- |
PREDICTED: serine carboxypeptidase-like 42 [Jatropha curcas] |
| 16 |
Hb_001674_010 |
0.0852756193 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 17 |
Hb_010962_020 |
0.0855885086 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_0815310 [Ricinus communis] |
| 18 |
Hb_098315_060 |
0.0864025863 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 19 |
Hb_010407_130 |
0.087277741 |
- |
- |
hypothetical protein POPTR_0008s16680g [Populus trichocarpa] |
| 20 |
Hb_004223_060 |
0.0875541169 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |