| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
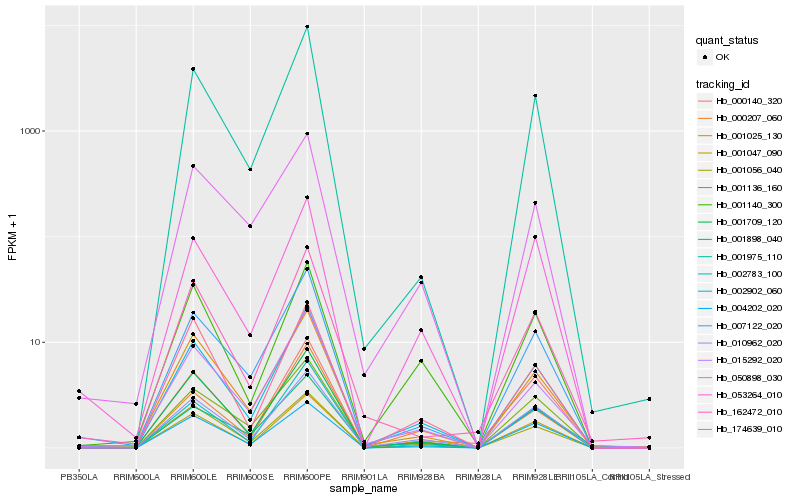
Hb_002902_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639824 [Jatropha curcas] |
| 2 |
Hb_010962_020 |
0.0557341522 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_0815310 [Ricinus communis] |
| 3 |
Hb_001047_090 |
0.0669786217 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP16-like [Jatropha curcas] |
| 4 |
Hb_001709_120 |
0.0761271903 |
- |
- |
hypothetical protein RCOM_1313640 [Ricinus communis] |
| 5 |
Hb_015292_020 |
0.0799118676 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_007122_020 |
0.0802770316 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 7 |
Hb_001975_110 |
0.082007072 |
- |
- |
- |
| 8 |
Hb_001025_130 |
0.0828782414 |
- |
- |
PREDICTED: uncharacterized protein LOC105640342 [Jatropha curcas] |
| 9 |
Hb_001136_160 |
0.0898776702 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_001056_040 |
0.0926549115 |
- |
- |
hypothetical protein POPTR_0006s04450g [Populus trichocarpa] |
| 11 |
Hb_001898_040 |
0.0983732282 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 12 |
Hb_001140_300 |
0.1011363595 |
- |
- |
PREDICTED: tubulin beta-1 chain [Cicer arietinum] |
| 13 |
Hb_050898_030 |
0.1030789827 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_174639_010 |
0.1040692499 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_053264_010 |
0.1058072744 |
- |
- |
beta glucosidase [Hevea brasiliensis] |
| 16 |
Hb_004202_020 |
0.10616101 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_162472_010 |
0.106907541 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002783_100 |
0.1071038015 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 19 |
Hb_000140_320 |
0.1084133234 |
- |
- |
delta-8 sphingolipid desaturase [Vernicia fordii] |
| 20 |
Hb_000207_060 |
0.1087232989 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |