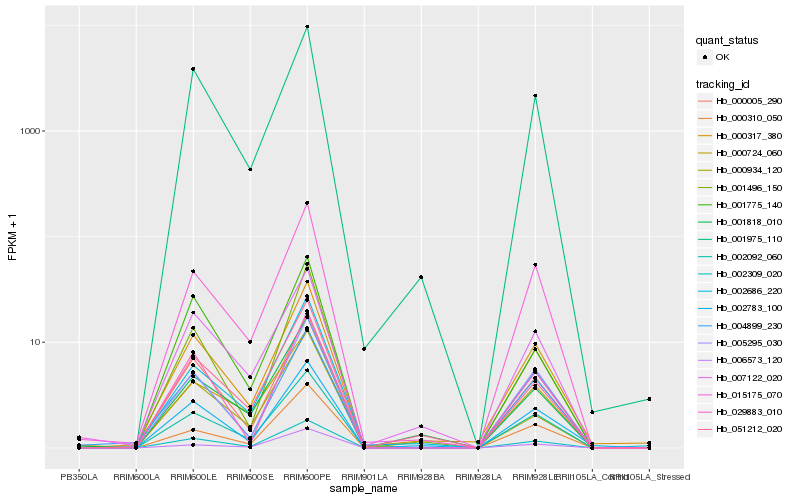
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002309_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_26057 [Jatropha curcas] |
| 2 |
Hb_002092_060 |
0.0440655201 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000005_290 |
0.0492394827 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 4 |
Hb_015175_070 |
0.0695845914 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 5 |
Hb_001975_110 |
0.0740348168 |
- |
- |
- |
| 6 |
Hb_005295_030 |
0.076909178 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |
| 7 |
Hb_001496_150 |
0.0787873711 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 8 |
Hb_000317_380 |
0.0795759035 |
- |
- |
Peroxidase 47 precursor, putative [Ricinus communis] |
| 9 |
Hb_002686_220 |
0.0824108375 |
- |
- |
PREDICTED: expansin-A15-like [Jatropha curcas] |
| 10 |
Hb_002783_100 |
0.0865883344 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 11 |
Hb_000310_050 |
0.0871735858 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 12 |
Hb_051212_020 |
0.0907783486 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2-like [Jatropha curcas] |
| 13 |
Hb_029883_010 |
0.0941610136 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 14 |
Hb_004899_230 |
0.0957768313 |
- |
- |
Indole-3-acetic acid-induced protein ARG7, putative [Ricinus communis] |
| 15 |
Hb_000724_060 |
0.0990925071 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 16 |
Hb_000934_120 |
0.1011392415 |
- |
- |
PREDICTED: tumor necrosis factor ligand superfamily member 6 [Populus euphratica] |
| 17 |
Hb_001775_140 |
0.1016678083 |
- |
- |
Phytosulfokine 3 precursor, putative isoform 2 [Theobroma cacao] |
| 18 |
Hb_006573_120 |
0.104594639 |
- |
- |
PREDICTED: DUF21 domain-containing protein At5g52790 [Jatropha curcas] |
| 19 |
Hb_007122_020 |
0.1064520638 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 20 |
Hb_001818_010 |
0.107009124 |
- |
- |
Disease resistance response protein, putative [Ricinus communis] |