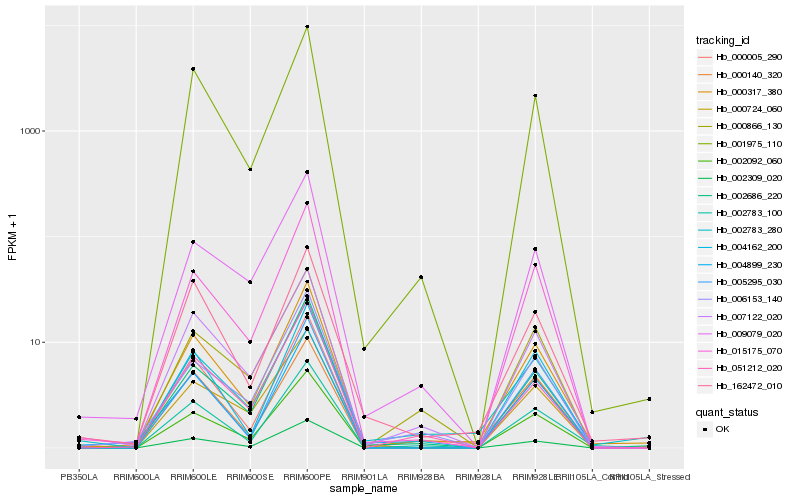
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000317_380 |
0.0 |
- |
- |
Peroxidase 47 precursor, putative [Ricinus communis] |
| 2 |
Hb_002783_100 |
0.0735582073 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 3 |
Hb_002092_060 |
0.077066393 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001975_110 |
0.078537205 |
- |
- |
- |
| 5 |
Hb_002309_020 |
0.0795759035 |
- |
- |
hypothetical protein JCGZ_26057 [Jatropha curcas] |
| 6 |
Hb_015175_070 |
0.0812938624 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 7 |
Hb_000005_290 |
0.0818329372 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 8 |
Hb_002783_280 |
0.0898459683 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 9 |
Hb_002686_220 |
0.0907454064 |
- |
- |
PREDICTED: expansin-A15-like [Jatropha curcas] |
| 10 |
Hb_007122_020 |
0.0942018941 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 11 |
Hb_005295_030 |
0.0992749734 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |
| 12 |
Hb_004899_230 |
0.1007846303 |
- |
- |
Indole-3-acetic acid-induced protein ARG7, putative [Ricinus communis] |
| 13 |
Hb_006153_140 |
0.1026929917 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 3 [Jatropha curcas] |
| 14 |
Hb_004162_200 |
0.1027066944 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |
| 15 |
Hb_162472_010 |
0.1043190336 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_051212_020 |
0.105370967 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2-like [Jatropha curcas] |
| 17 |
Hb_000866_130 |
0.1076300547 |
- |
- |
hypothetical protein JCGZ_14397 [Jatropha curcas] |
| 18 |
Hb_000140_320 |
0.1083090958 |
- |
- |
delta-8 sphingolipid desaturase [Vernicia fordii] |
| 19 |
Hb_009079_020 |
0.1092864406 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 20 |
Hb_000724_060 |
0.1107231161 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |