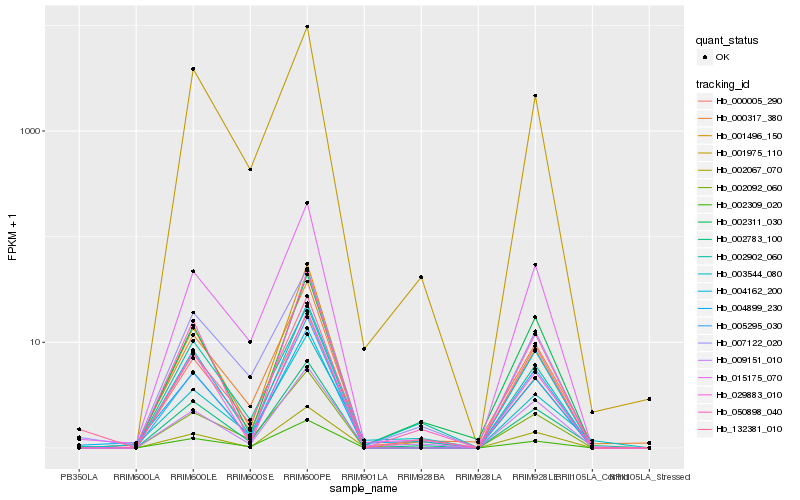
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002783_100 |
0.0 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 2 |
Hb_000317_380 |
0.0735582073 |
- |
- |
Peroxidase 47 precursor, putative [Ricinus communis] |
| 3 |
Hb_000005_290 |
0.0817405409 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 4 |
Hb_001975_110 |
0.0818022576 |
- |
- |
- |
| 5 |
Hb_050898_040 |
0.0865593986 |
- |
- |
- |
| 6 |
Hb_002309_020 |
0.0865883344 |
- |
- |
hypothetical protein JCGZ_26057 [Jatropha curcas] |
| 7 |
Hb_004162_200 |
0.0878384226 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |
| 8 |
Hb_005295_030 |
0.0905281412 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |
| 9 |
Hb_002092_060 |
0.0910163288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_029883_010 |
0.1001736765 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 11 |
Hb_015175_070 |
0.1009690709 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 12 |
Hb_009151_010 |
0.1041707239 |
- |
- |
Peroxidase 3 [Morus notabilis] |
| 13 |
Hb_004899_230 |
0.1045011793 |
- |
- |
Indole-3-acetic acid-induced protein ARG7, putative [Ricinus communis] |
| 14 |
Hb_001496_150 |
0.1057923071 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 15 |
Hb_002902_060 |
0.1071038015 |
- |
- |
PREDICTED: uncharacterized protein LOC105639824 [Jatropha curcas] |
| 16 |
Hb_007122_020 |
0.1077454663 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 17 |
Hb_002067_070 |
0.1096070744 |
- |
- |
hypothetical protein JCGZ_19254 [Jatropha curcas] |
| 18 |
Hb_003544_080 |
0.1100452762 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 19 |
Hb_132381_010 |
0.1100969809 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 20 |
Hb_002311_030 |
0.113267307 |
- |
- |
conserved hypothetical protein [Ricinus communis] |