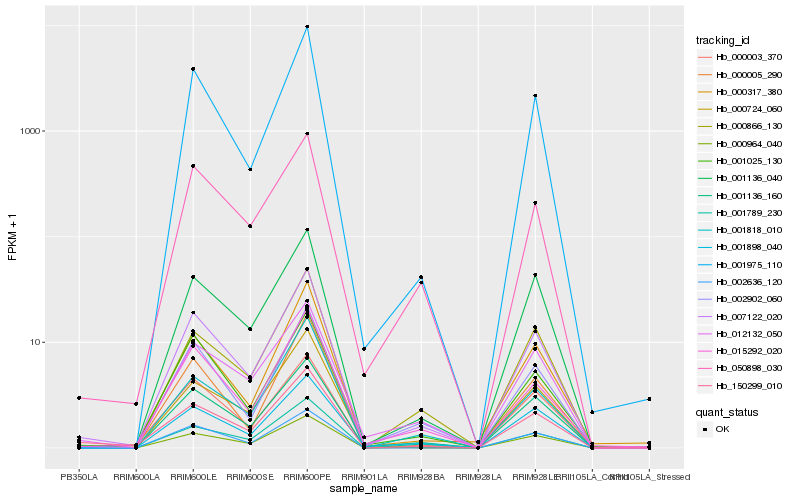
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007122_020 |
0.0 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 2 |
Hb_001975_110 |
0.0696597303 |
- |
- |
- |
| 3 |
Hb_001136_160 |
0.0723417619 |
- |
- |
catalytic, putative [Ricinus communis] |
| 4 |
Hb_000964_040 |
0.0732844787 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER isoform X2 [Jatropha curcas] |
| 5 |
Hb_150299_010 |
0.0801299277 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 1-like [Tarenaya hassleriana] |
| 6 |
Hb_002902_060 |
0.0802770316 |
- |
- |
PREDICTED: uncharacterized protein LOC105639824 [Jatropha curcas] |
| 7 |
Hb_001136_040 |
0.0850927758 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_015292_020 |
0.087004687 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_001898_040 |
0.0898513784 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000003_370 |
0.0912759155 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 11 |
Hb_050898_030 |
0.0914902263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000724_060 |
0.0920851549 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 13 |
Hb_002636_120 |
0.0926839515 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 14 |
Hb_001025_130 |
0.0928534425 |
- |
- |
PREDICTED: uncharacterized protein LOC105640342 [Jatropha curcas] |
| 15 |
Hb_000317_380 |
0.0942018941 |
- |
- |
Peroxidase 47 precursor, putative [Ricinus communis] |
| 16 |
Hb_001818_010 |
0.0942164405 |
- |
- |
Disease resistance response protein, putative [Ricinus communis] |
| 17 |
Hb_001789_230 |
0.0948617468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000005_290 |
0.0953102464 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 19 |
Hb_000866_130 |
0.0955519984 |
- |
- |
hypothetical protein JCGZ_14397 [Jatropha curcas] |
| 20 |
Hb_012132_050 |
0.0960538177 |
- |
- |
PREDICTED: aspartic proteinase PCS1 [Jatropha curcas] |