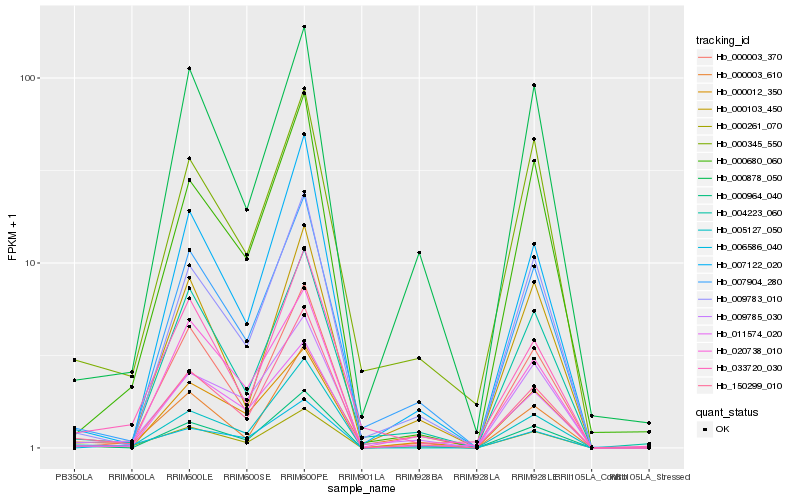
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_070 |
0.0 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g06240-like [Jatropha curcas] |
| 2 |
Hb_150299_010 |
0.0957040775 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 1-like [Tarenaya hassleriana] |
| 3 |
Hb_000003_370 |
0.1042672031 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 4 |
Hb_007904_280 |
0.1072899952 |
- |
- |
PREDICTED: uncharacterized protein LOC105644987 [Jatropha curcas] |
| 5 |
Hb_000964_040 |
0.110631216 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER isoform X2 [Jatropha curcas] |
| 6 |
Hb_009785_030 |
0.1132271645 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |
| 7 |
Hb_004223_060 |
0.1153572175 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 8 |
Hb_000012_350 |
0.1177699267 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein ATH1 [Jatropha curcas] |
| 9 |
Hb_000878_050 |
0.1184260263 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 10 |
Hb_000345_550 |
0.1193064347 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 11 |
Hb_006586_040 |
0.1211235361 |
- |
- |
hypothetical protein POPTR_0011s11750g [Populus trichocarpa] |
| 12 |
Hb_009783_010 |
0.121892807 |
- |
- |
PREDICTED: sulfated surface glycoprotein 185-like [Populus euphratica] |
| 13 |
Hb_000003_610 |
0.1247099636 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase [Jatropha curcas] |
| 14 |
Hb_005127_050 |
0.1279465735 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA2-like [Populus euphratica] |
| 15 |
Hb_033720_030 |
0.1281504019 |
- |
- |
KDEL motif-containing protein 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_011574_020 |
0.1297126023 |
- |
- |
hypothetical protein JCGZ_21524 [Jatropha curcas] |
| 17 |
Hb_007122_020 |
0.1303041536 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 18 |
Hb_020738_010 |
0.1318648052 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At2g24230 [Jatropha curcas] |
| 19 |
Hb_000103_450 |
0.1322383237 |
- |
- |
PREDICTED: dentin sialophosphoprotein-like [Jatropha curcas] |
| 20 |
Hb_000680_060 |
0.1348481616 |
- |
- |
hypothetical protein POPTR_0001s20790g [Populus trichocarpa] |