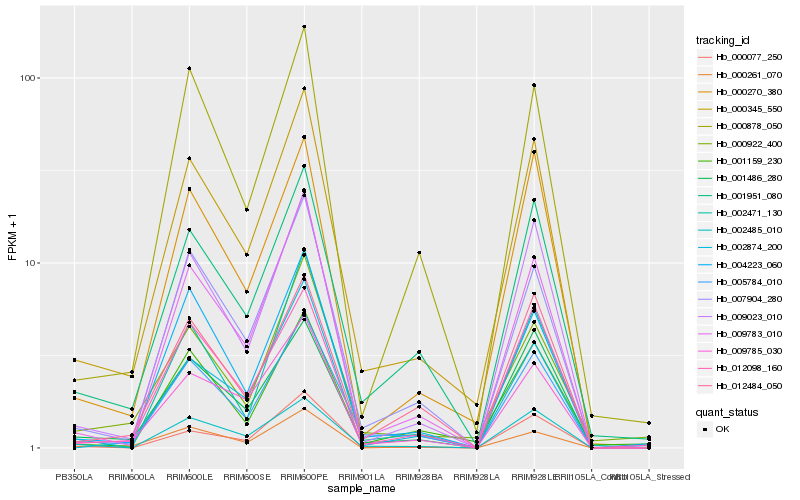
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000345_550 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 2 |
Hb_005784_010 |
0.0799217715 |
- |
- |
PREDICTED: uncharacterized protein LOC105645175 [Jatropha curcas] |
| 3 |
Hb_009785_030 |
0.086661905 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |
| 4 |
Hb_001951_080 |
0.088279482 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 5 |
Hb_009783_010 |
0.0901915824 |
- |
- |
PREDICTED: sulfated surface glycoprotein 185-like [Populus euphratica] |
| 6 |
Hb_009023_010 |
0.0936991984 |
- |
- |
glycosyl hydrolase family 31 family protein [Populus trichocarpa] |
| 7 |
Hb_007904_280 |
0.0955835147 |
- |
- |
PREDICTED: uncharacterized protein LOC105644987 [Jatropha curcas] |
| 8 |
Hb_001486_280 |
0.0971635648 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |
| 9 |
Hb_000270_380 |
0.0989336757 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 10 |
Hb_002485_010 |
0.106615476 |
- |
- |
PREDICTED: extensin [Jatropha curcas] |
| 11 |
Hb_012098_160 |
0.1080951318 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 12 |
Hb_001159_230 |
0.1112320183 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 13 |
Hb_004223_060 |
0.1121562552 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 14 |
Hb_012484_050 |
0.1137840275 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 15 |
Hb_000922_400 |
0.1138550497 |
- |
- |
PREDICTED: uncharacterized protein LOC105640363 [Jatropha curcas] |
| 16 |
Hb_000878_050 |
0.118082002 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 17 |
Hb_002874_200 |
0.1182055911 |
- |
- |
PREDICTED: uncharacterized protein LOC105109832 [Populus euphratica] |
| 18 |
Hb_000261_070 |
0.1193064347 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g06240-like [Jatropha curcas] |
| 19 |
Hb_002471_130 |
0.1212602456 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 3 [Jatropha curcas] |
| 20 |
Hb_000077_250 |
0.12216028 |
- |
- |
hypothetical protein JCGZ_25802 [Jatropha curcas] |