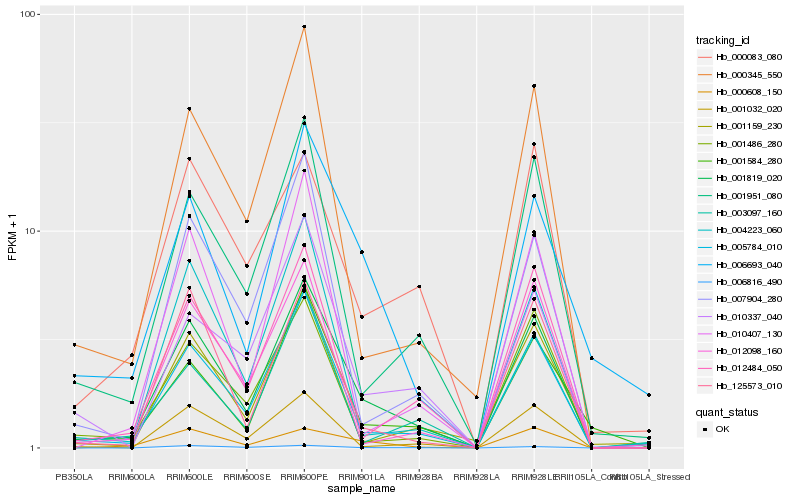
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001819_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641890 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005784_010 |
0.0923854734 |
- |
- |
PREDICTED: uncharacterized protein LOC105645175 [Jatropha curcas] |
| 3 |
Hb_001951_080 |
0.1354306862 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 4 |
Hb_000345_550 |
0.1473517411 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 5 |
Hb_012484_050 |
0.1511599696 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 6 |
Hb_004223_060 |
0.1535985957 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 7 |
Hb_001032_020 |
0.1561513092 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 8 |
Hb_006693_040 |
0.1580666803 |
- |
- |
PREDICTED: homeotic protein female sterile-like [Jatropha curcas] |
| 9 |
Hb_000608_150 |
0.1587845853 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001159_230 |
0.1592501387 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 11 |
Hb_001584_280 |
0.1600029521 |
- |
- |
PREDICTED: uncharacterized protein LOC105647395 [Jatropha curcas] |
| 12 |
Hb_125573_010 |
0.1603424926 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 13 |
Hb_001486_280 |
0.161096616 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |
| 14 |
Hb_007904_280 |
0.1620573083 |
- |
- |
PREDICTED: uncharacterized protein LOC105644987 [Jatropha curcas] |
| 15 |
Hb_003097_160 |
0.16214823 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein sel-10 [Jatropha curcas] |
| 16 |
Hb_012098_160 |
0.1625975664 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 17 |
Hb_000083_080 |
0.1633269676 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 18 |
Hb_006816_490 |
0.1646162846 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 21 [Jatropha curcas] |
| 19 |
Hb_010407_130 |
0.1665861911 |
- |
- |
hypothetical protein POPTR_0008s16680g [Populus trichocarpa] |
| 20 |
Hb_010337_040 |
0.1674754574 |
- |
- |
- |