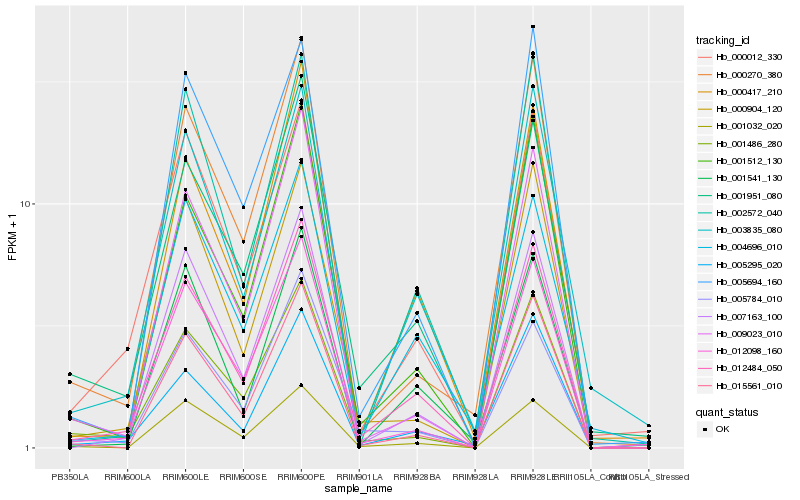
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012484_050 |
0.0 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 2 |
Hb_001951_080 |
0.0795587476 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 3 |
Hb_007163_100 |
0.0835704553 |
- |
- |
BnaC03g52280D [Brassica napus] |
| 4 |
Hb_012098_160 |
0.0849310604 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 5 |
Hb_001512_130 |
0.090374818 |
- |
- |
PREDICTED: uncharacterized protein LOC105633633 [Jatropha curcas] |
| 6 |
Hb_000012_330 |
0.0910219907 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 7 |
Hb_001486_280 |
0.0920353171 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |
| 8 |
Hb_000417_210 |
0.0925760935 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_015561_010 |
0.0969346906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001032_020 |
0.097282591 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 11 |
Hb_005295_020 |
0.0985731865 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 12 |
Hb_003835_080 |
0.1027957497 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000270_380 |
0.1035213516 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 14 |
Hb_001541_130 |
0.1036909883 |
- |
- |
GDSL esterase/lipase [Morus notabilis] |
| 15 |
Hb_000904_120 |
0.1037931796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004696_010 |
0.1056244506 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 17 |
Hb_009023_010 |
0.1068632945 |
- |
- |
glycosyl hydrolase family 31 family protein [Populus trichocarpa] |
| 18 |
Hb_002572_040 |
0.107475022 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 19 |
Hb_005784_010 |
0.1096216973 |
- |
- |
PREDICTED: uncharacterized protein LOC105645175 [Jatropha curcas] |
| 20 |
Hb_005694_160 |
0.1098536422 |
- |
- |
conserved hypothetical protein [Ricinus communis] |