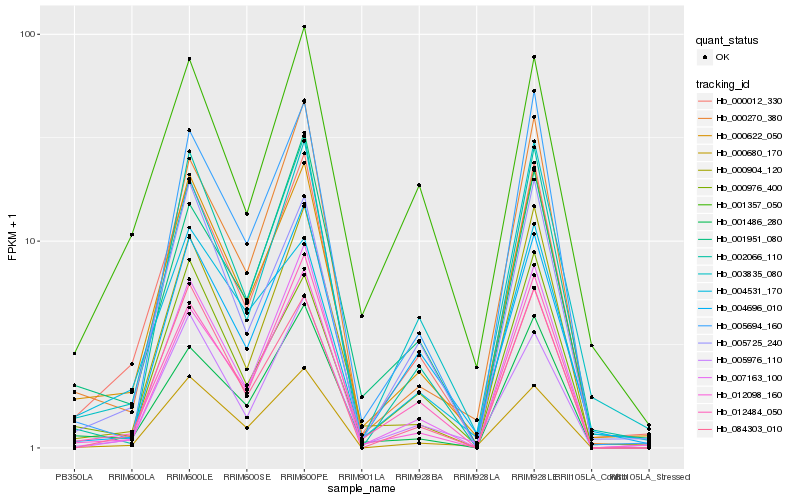
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000012_330 |
0.0 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 2 |
Hb_000622_050 |
0.0541779045 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_012098_160 |
0.0873880204 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 4 |
Hb_003835_080 |
0.089198894 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_012484_050 |
0.0910219907 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 6 |
Hb_005725_240 |
0.0964516126 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH74 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001486_280 |
0.1040621153 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |
| 8 |
Hb_007163_100 |
0.1043851731 |
- |
- |
BnaC03g52280D [Brassica napus] |
| 9 |
Hb_002066_110 |
0.1104178024 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 10 |
Hb_000904_120 |
0.1105648621 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000270_380 |
0.111958216 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 12 |
Hb_000976_400 |
0.1128786607 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 13 |
Hb_005976_110 |
0.1129481942 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 14 |
Hb_005694_160 |
0.1145088065 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004531_170 |
0.1145184879 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 16 |
Hb_001951_080 |
0.1150014186 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 17 |
Hb_004696_010 |
0.1151696668 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 18 |
Hb_000680_170 |
0.1157533133 |
- |
- |
hypothetical protein JCGZ_24789 [Jatropha curcas] |
| 19 |
Hb_001357_050 |
0.1159890386 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12-like [Jatropha curcas] |
| 20 |
Hb_084303_010 |
0.1172675864 |
- |
- |
conserved hypothetical protein [Ricinus communis] |