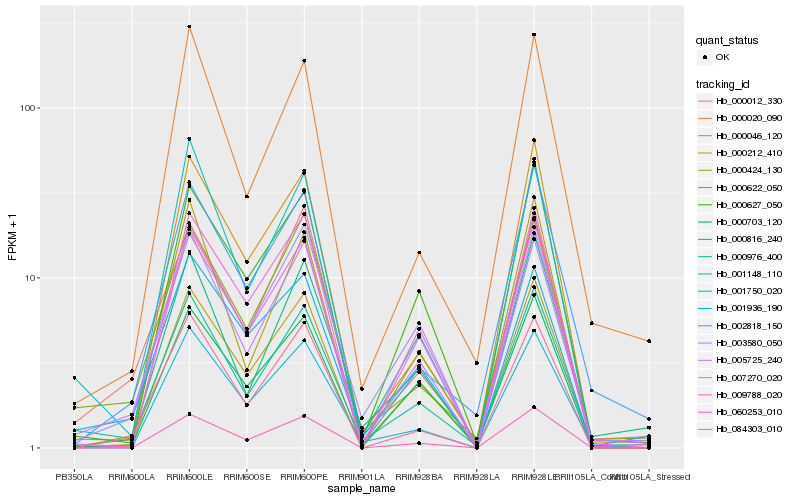
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005725_240 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH74 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000976_400 |
0.059503513 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 3 |
Hb_000212_410 |
0.078436382 |
- |
- |
PREDICTED: serine carboxypeptidase 1-like [Jatropha curcas] |
| 4 |
Hb_084303_010 |
0.088875921 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000622_050 |
0.0925063255 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000012_330 |
0.0964516126 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 7 |
Hb_000816_240 |
0.0981153665 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_009788_020 |
0.0990727969 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g06840 [Jatropha curcas] |
| 9 |
Hb_001936_190 |
0.1022116563 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 10 |
Hb_007270_020 |
0.1030995533 |
- |
- |
hypothetical protein JCGZ_09593 [Jatropha curcas] |
| 11 |
Hb_000627_050 |
0.1041542421 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 12 |
Hb_000020_090 |
0.1042470764 |
- |
- |
hypothetical protein JCGZ_09084 [Jatropha curcas] |
| 13 |
Hb_001148_110 |
0.1052097668 |
- |
- |
- |
| 14 |
Hb_060253_010 |
0.1052119033 |
- |
- |
hypothetical protein JCGZ_14707 [Jatropha curcas] |
| 15 |
Hb_000424_130 |
0.1067103784 |
- |
- |
PREDICTED: monoacylglycerol lipase abhd6-B-like [Jatropha curcas] |
| 16 |
Hb_000703_120 |
0.1080092027 |
- |
- |
PREDICTED: uncharacterized protein LOC105635706 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001750_020 |
0.1092011639 |
- |
- |
PREDICTED: MATE efflux family protein 6-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_003580_050 |
0.1093429026 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 19 |
Hb_002818_150 |
0.1097286228 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0014s10680g [Populus trichocarpa] |
| 20 |
Hb_000046_120 |
0.1103264272 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |