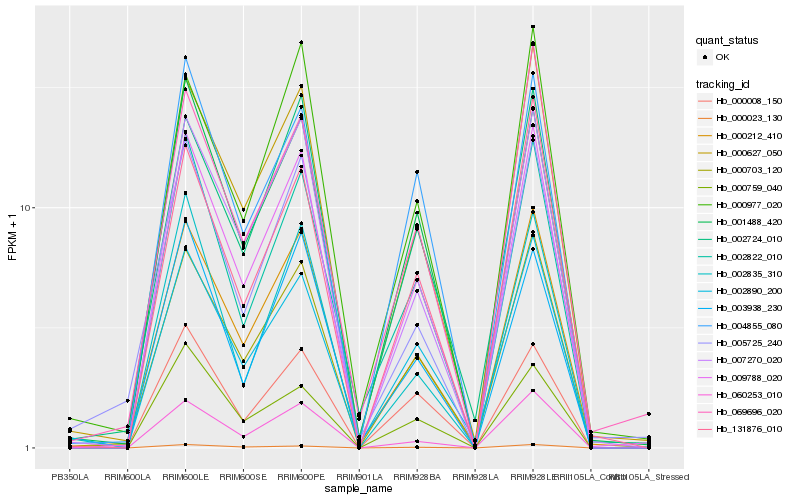
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009788_020 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g06840 [Jatropha curcas] |
| 2 |
Hb_000212_410 |
0.0481136541 |
- |
- |
PREDICTED: serine carboxypeptidase 1-like [Jatropha curcas] |
| 3 |
Hb_000703_120 |
0.0488483745 |
- |
- |
PREDICTED: uncharacterized protein LOC105635706 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000627_050 |
0.0614327244 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 5 |
Hb_007270_020 |
0.0635756059 |
- |
- |
hypothetical protein JCGZ_09593 [Jatropha curcas] |
| 6 |
Hb_060253_010 |
0.0742210305 |
- |
- |
hypothetical protein JCGZ_14707 [Jatropha curcas] |
| 7 |
Hb_001488_420 |
0.0823284605 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 8 |
Hb_000023_130 |
0.0836154771 |
- |
- |
Metal-nicotianamine transporter YSL1 [Glycine soja] |
| 9 |
Hb_002724_010 |
0.0842005762 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Citrus sinensis] |
| 10 |
Hb_002890_200 |
0.0865717517 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 11 |
Hb_002822_010 |
0.0875144912 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Populus euphratica] |
| 12 |
Hb_004855_080 |
0.0911292044 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 13 |
Hb_000008_150 |
0.0954655197 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 14 |
Hb_131876_010 |
0.0970658441 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_002835_310 |
0.0977121228 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_003938_230 |
0.0984091754 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 17 |
Hb_005725_240 |
0.0990727969 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH74 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000977_020 |
0.0991979942 |
- |
- |
beta-hexosaminidase, putative [Ricinus communis] |
| 19 |
Hb_069696_020 |
0.1011184253 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000759_040 |
0.1017364456 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |