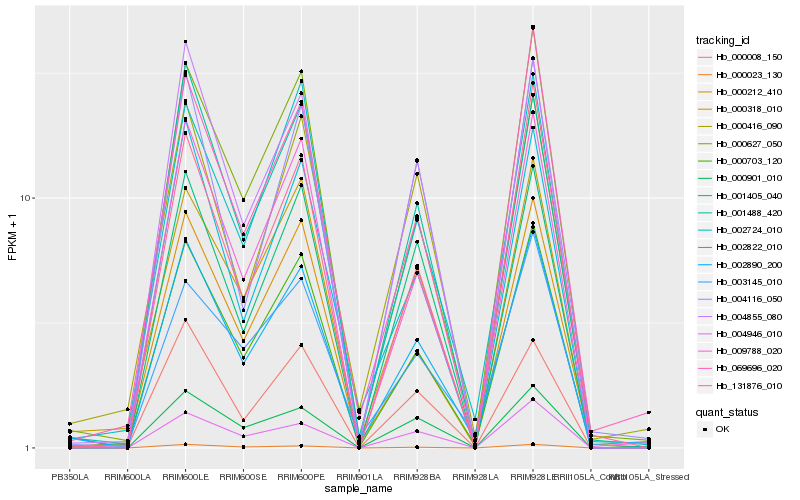
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002890_200 |
0.0 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 2 |
Hb_000703_120 |
0.0668564188 |
- |
- |
PREDICTED: uncharacterized protein LOC105635706 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000627_050 |
0.0816613103 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 4 |
Hb_009788_020 |
0.0865717517 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g06840 [Jatropha curcas] |
| 5 |
Hb_002822_010 |
0.0887363817 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Populus euphratica] |
| 6 |
Hb_004855_080 |
0.0890127052 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 7 |
Hb_001488_420 |
0.0933490997 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 8 |
Hb_000212_410 |
0.0939594992 |
- |
- |
PREDICTED: serine carboxypeptidase 1-like [Jatropha curcas] |
| 9 |
Hb_000318_010 |
0.0949534908 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 10 |
Hb_002724_010 |
0.0970916115 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Citrus sinensis] |
| 11 |
Hb_000023_130 |
0.1024787339 |
- |
- |
Metal-nicotianamine transporter YSL1 [Glycine soja] |
| 12 |
Hb_004946_010 |
0.105816057 |
- |
- |
hypothetical protein POPTR_0013s035502g, partial [Populus trichocarpa] |
| 13 |
Hb_069696_020 |
0.1058546125 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_001405_040 |
0.1068609834 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 15 |
Hb_003145_010 |
0.110023691 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 16 |
Hb_131876_010 |
0.1102762297 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_004116_050 |
0.11110776 |
- |
- |
PREDICTED: cytochrome P450 90A1 [Jatropha curcas] |
| 18 |
Hb_000416_090 |
0.1135414124 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 19 |
Hb_000008_150 |
0.1147004018 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 20 |
Hb_000901_010 |
0.1163356423 |
- |
- |
PREDICTED: putative G-type lectin S-receptor-like serine/threonine-protein kinase At1g61610 [Jatropha curcas] |