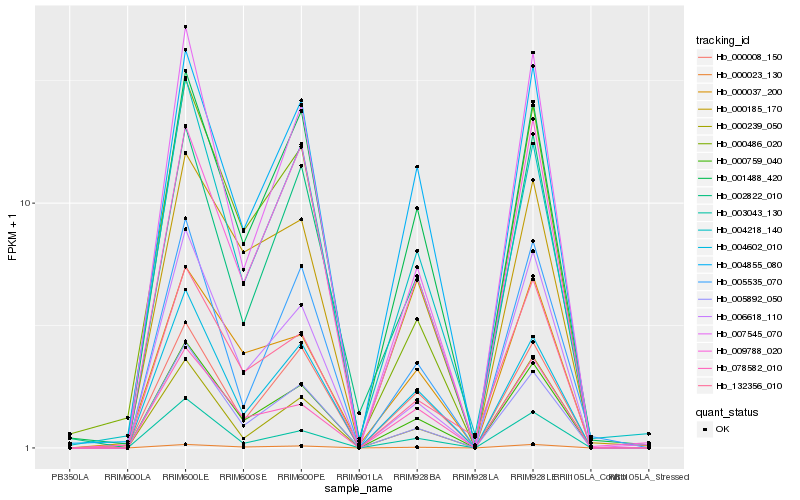
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000759_040 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 2 |
Hb_005892_050 |
0.0505949641 |
- |
- |
PREDICTED: uncharacterized protein LOC105632175 [Jatropha curcas] |
| 3 |
Hb_004602_010 |
0.069884574 |
- |
- |
PREDICTED: uncharacterized protein LOC105042236 isoform X2 [Elaeis guineensis] |
| 4 |
Hb_000023_130 |
0.0706605466 |
- |
- |
Metal-nicotianamine transporter YSL1 [Glycine soja] |
| 5 |
Hb_004855_080 |
0.073174086 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 6 |
Hb_001488_420 |
0.0769021152 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 7 |
Hb_006618_110 |
0.0859033975 |
- |
- |
- |
| 8 |
Hb_000008_150 |
0.0925374848 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 9 |
Hb_078582_010 |
0.0928998255 |
- |
- |
hypothetical protein POPTR_0005s01620g [Populus trichocarpa] |
| 10 |
Hb_003043_130 |
0.0941480345 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF2-like [Jatropha curcas] |
| 11 |
Hb_004218_140 |
0.0945729015 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 isoform X3 [Jatropha curcas] |
| 12 |
Hb_000185_170 |
0.0958148159 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_132356_010 |
0.0961355333 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 14 |
Hb_007545_070 |
0.0990173655 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 306 [Populus euphratica] |
| 15 |
Hb_009788_020 |
0.1017364456 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g06840 [Jatropha curcas] |
| 16 |
Hb_000037_200 |
0.1024521298 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_002822_010 |
0.1044877689 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Populus euphratica] |
| 18 |
Hb_000486_020 |
0.1061261631 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.2 [Jatropha curcas] |
| 19 |
Hb_005535_070 |
0.1129954082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000239_050 |
0.1166106545 |
- |
- |
PREDICTED: cyclin-D5-1-like [Jatropha curcas] |