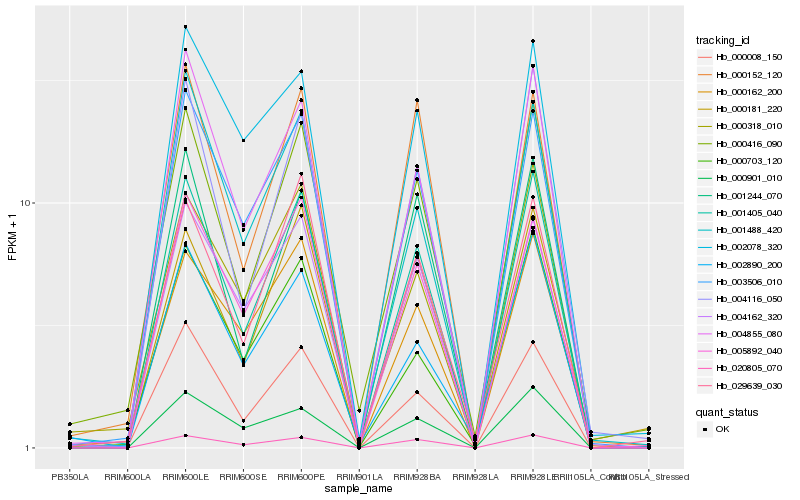
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001405_040 |
0.0 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 2 |
Hb_020805_070 |
0.0637823898 |
- |
- |
PREDICTED: MLO protein homolog 1-like [Jatropha curcas] |
| 3 |
Hb_004855_080 |
0.0712460449 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 4 |
Hb_000008_150 |
0.0745511504 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 5 |
Hb_004116_050 |
0.0751568136 |
- |
- |
PREDICTED: cytochrome P450 90A1 [Jatropha curcas] |
| 6 |
Hb_003506_010 |
0.0771024975 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 7 |
Hb_005892_040 |
0.0831016232 |
- |
- |
hypothetical protein JCGZ_02368 [Jatropha curcas] |
| 8 |
Hb_004162_320 |
0.0888413527 |
- |
- |
PREDICTED: uncharacterized protein LOC105628009 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001244_070 |
0.0922565901 |
- |
- |
hypothetical protein POPTR_0004s13470g [Populus trichocarpa] |
| 10 |
Hb_000416_090 |
0.0934085782 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 11 |
Hb_029639_030 |
0.0947808011 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 2-like [Jatropha curcas] |
| 12 |
Hb_000162_200 |
0.0948585932 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001488_420 |
0.0974435997 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 14 |
Hb_000181_220 |
0.1011803889 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000318_010 |
0.1026443654 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 16 |
Hb_000901_010 |
0.1032210695 |
- |
- |
PREDICTED: putative G-type lectin S-receptor-like serine/threonine-protein kinase At1g61610 [Jatropha curcas] |
| 17 |
Hb_002078_320 |
0.1063555835 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 18 |
Hb_000703_120 |
0.1065559594 |
- |
- |
PREDICTED: uncharacterized protein LOC105635706 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002890_200 |
0.1068609834 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 20 |
Hb_000152_120 |
0.107425879 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |