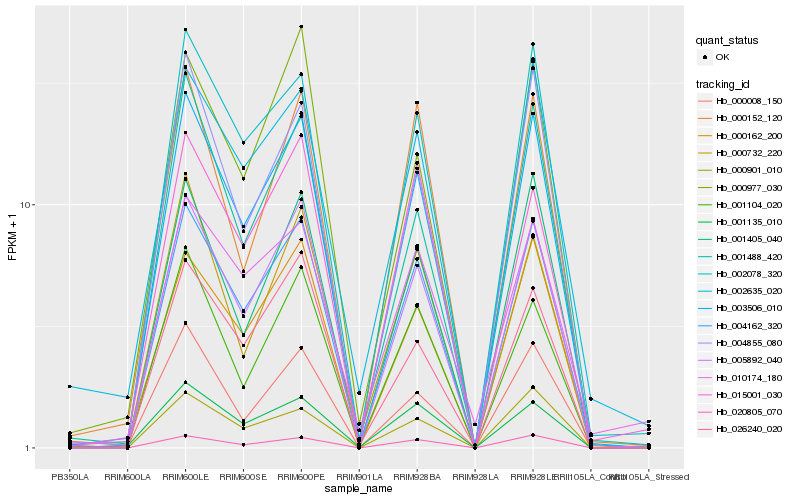
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004162_320 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628009 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003506_010 |
0.0399593975 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 3 |
Hb_005892_040 |
0.057314559 |
- |
- |
hypothetical protein JCGZ_02368 [Jatropha curcas] |
| 4 |
Hb_020805_070 |
0.0663679118 |
- |
- |
PREDICTED: MLO protein homolog 1-like [Jatropha curcas] |
| 5 |
Hb_002078_320 |
0.0694402009 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 6 |
Hb_000162_200 |
0.0817381958 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001405_040 |
0.0888413527 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 8 |
Hb_001135_010 |
0.0894846349 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 9 |
Hb_026240_020 |
0.0946086861 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 10 |
Hb_001104_020 |
0.0968995836 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_002635_020 |
0.097857969 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 12 |
Hb_015001_030 |
0.1039921151 |
- |
- |
inosine-uridine preferring nucleoside hydrolase family protein [Populus trichocarpa] |
| 13 |
Hb_004855_080 |
0.1046160058 |
- |
- |
hypothetical protein JCGZ_25039 [Jatropha curcas] |
| 14 |
Hb_000152_120 |
0.1064688452 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |
| 15 |
Hb_001488_420 |
0.1068012637 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 16 |
Hb_000977_030 |
0.1106731577 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_000901_010 |
0.1112912183 |
- |
- |
PREDICTED: putative G-type lectin S-receptor-like serine/threonine-protein kinase At1g61610 [Jatropha curcas] |
| 18 |
Hb_000008_150 |
0.1114806083 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 19 |
Hb_000732_220 |
0.1127395145 |
- |
- |
PREDICTED: importin subunit alpha-5 [Jatropha curcas] |
| 20 |
Hb_010174_180 |
0.1139087743 |
- |
- |
protein binding protein, putative [Ricinus communis] |