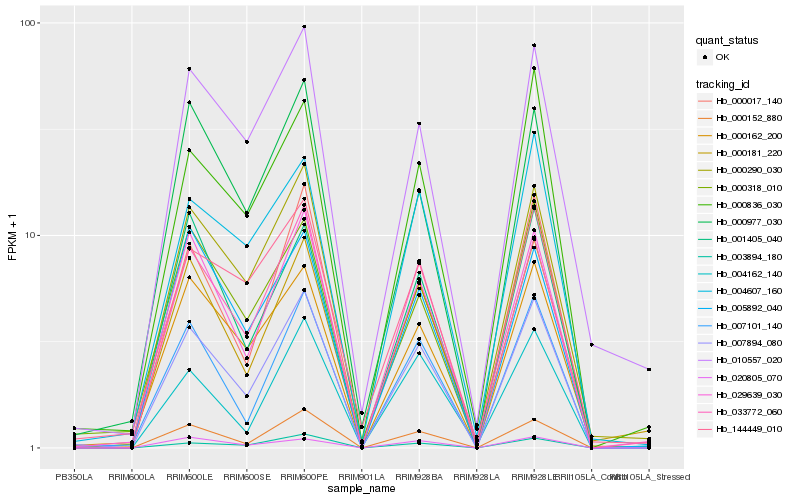
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007894_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000152_880 |
0.0750181538 |
- |
- |
- |
| 3 |
Hb_033772_060 |
0.0759399911 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000017_140 |
0.0781242449 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 5 |
Hb_007101_140 |
0.0826691998 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 6 |
Hb_029639_030 |
0.0887055053 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 2-like [Jatropha curcas] |
| 7 |
Hb_000162_200 |
0.0912027745 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003894_180 |
0.0951209482 |
- |
- |
PREDICTED: uncharacterized protein LOC104245193, partial [Nicotiana sylvestris] |
| 9 |
Hb_000181_220 |
0.0982856336 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_000836_030 |
0.1083589585 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 11 |
Hb_001405_040 |
0.1088497112 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 12 |
Hb_000318_010 |
0.1095741639 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 13 |
Hb_004607_160 |
0.1126608886 |
- |
- |
PREDICTED: uncharacterized protein LOC105134931 [Populus euphratica] |
| 14 |
Hb_000977_030 |
0.1127762724 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_020805_070 |
0.1159291585 |
- |
- |
PREDICTED: MLO protein homolog 1-like [Jatropha curcas] |
| 16 |
Hb_010557_020 |
0.116383969 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 17 |
Hb_144449_010 |
0.119132757 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 18 |
Hb_005892_040 |
0.1192922163 |
- |
- |
hypothetical protein JCGZ_02368 [Jatropha curcas] |
| 19 |
Hb_004162_140 |
0.1207805695 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 20 |
Hb_000290_030 |
0.1213085783 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |