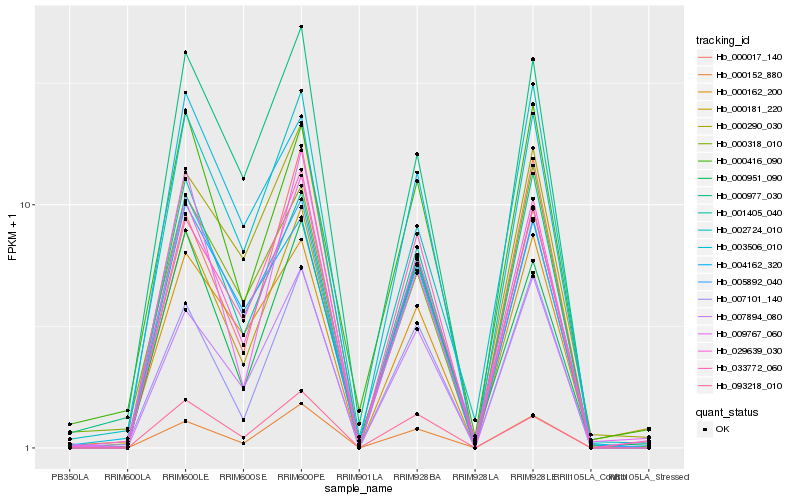
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029639_030 |
0.0 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 2-like [Jatropha curcas] |
| 2 |
Hb_000181_220 |
0.0736738612 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_000152_880 |
0.0820879897 |
- |
- |
- |
| 4 |
Hb_033772_060 |
0.0837351259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007894_080 |
0.0887055053 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005892_040 |
0.0908970616 |
- |
- |
hypothetical protein JCGZ_02368 [Jatropha curcas] |
| 7 |
Hb_000977_030 |
0.0921856126 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_007101_140 |
0.0927790996 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 9 |
Hb_000951_090 |
0.0934901296 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 10 |
Hb_001405_040 |
0.0947808011 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 11 |
Hb_000416_090 |
0.1002350339 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 12 |
Hb_000162_200 |
0.1020550055 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002724_010 |
0.1062176038 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Citrus sinensis] |
| 14 |
Hb_003506_010 |
0.1108349541 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 15 |
Hb_000017_140 |
0.1109029162 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 16 |
Hb_009767_060 |
0.1116305121 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 17 |
Hb_093218_010 |
0.111698852 |
- |
- |
hypothetical protein POPTR_0001s05220g [Populus trichocarpa] |
| 18 |
Hb_000290_030 |
0.1126882757 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 19 |
Hb_004162_320 |
0.1147727986 |
- |
- |
PREDICTED: uncharacterized protein LOC105628009 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000318_010 |
0.1151301104 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |