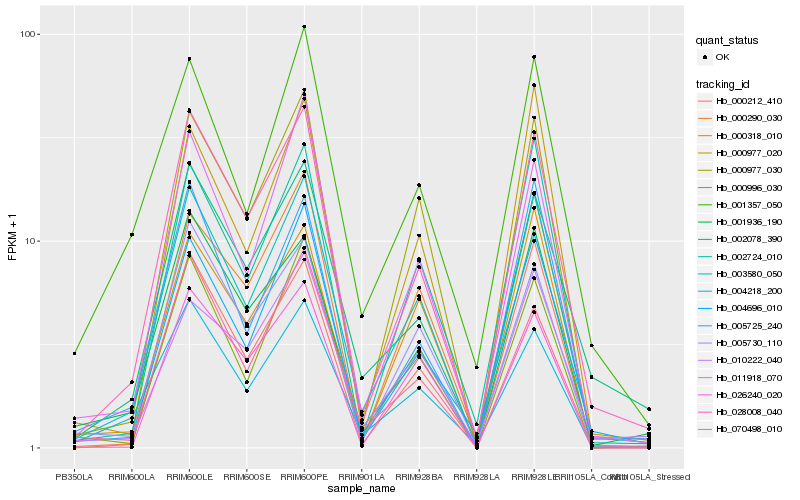
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003580_050 |
0.0 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 2 |
Hb_028008_040 |
0.0782080371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000977_030 |
0.081798784 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_000996_030 |
0.0844286926 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g63430 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004696_010 |
0.0920299589 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 6 |
Hb_002724_010 |
0.0957444974 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Citrus sinensis] |
| 7 |
Hb_004218_200 |
0.1010286867 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 8 |
Hb_000212_410 |
0.1024805242 |
- |
- |
PREDICTED: serine carboxypeptidase 1-like [Jatropha curcas] |
| 9 |
Hb_011918_070 |
0.1034661432 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 10 |
Hb_000318_010 |
0.1061848056 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 11 |
Hb_001936_190 |
0.1073650117 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 12 |
Hb_005730_110 |
0.1077192143 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 13 |
Hb_000290_030 |
0.1084768312 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 14 |
Hb_005725_240 |
0.1093429026 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH74 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000977_020 |
0.1108358599 |
- |
- |
beta-hexosaminidase, putative [Ricinus communis] |
| 16 |
Hb_010222_040 |
0.1124915662 |
- |
- |
PREDICTED: kinesin-2 [Jatropha curcas] |
| 17 |
Hb_070498_010 |
0.1136394558 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 18 |
Hb_026240_020 |
0.1149103071 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 19 |
Hb_002078_390 |
0.1150077506 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 20 |
Hb_001357_050 |
0.1158730461 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12-like [Jatropha curcas] |