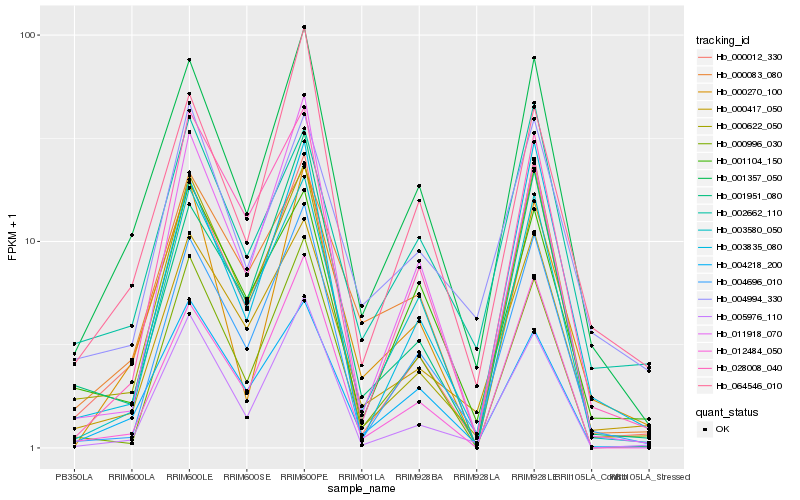
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001357_050 |
0.0 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12-like [Jatropha curcas] |
| 2 |
Hb_004218_200 |
0.1024332558 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 3 |
Hb_064546_010 |
0.1118143466 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 4 |
Hb_000417_050 |
0.1137200594 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 5 |
Hb_003580_050 |
0.1158730461 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 6 |
Hb_004696_010 |
0.1159443007 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 7 |
Hb_000012_330 |
0.1159890386 |
- |
- |
pollen specific protein sf21, putative [Ricinus communis] |
| 8 |
Hb_000270_100 |
0.1265043328 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 9 |
Hb_003835_080 |
0.1314177126 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_012484_050 |
0.1333020551 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 11 |
Hb_011918_070 |
0.1334300556 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 12 |
Hb_000622_050 |
0.135137685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000083_080 |
0.1352617338 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 14 |
Hb_001951_080 |
0.1360640875 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 15 |
Hb_001104_150 |
0.1376412576 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 16 |
Hb_002662_110 |
0.1410670022 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 17 |
Hb_005976_110 |
0.1412156644 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 18 |
Hb_004994_330 |
0.1416594351 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_028008_040 |
0.141693095 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000996_030 |
0.1418383736 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g63430 isoform X1 [Jatropha curcas] |