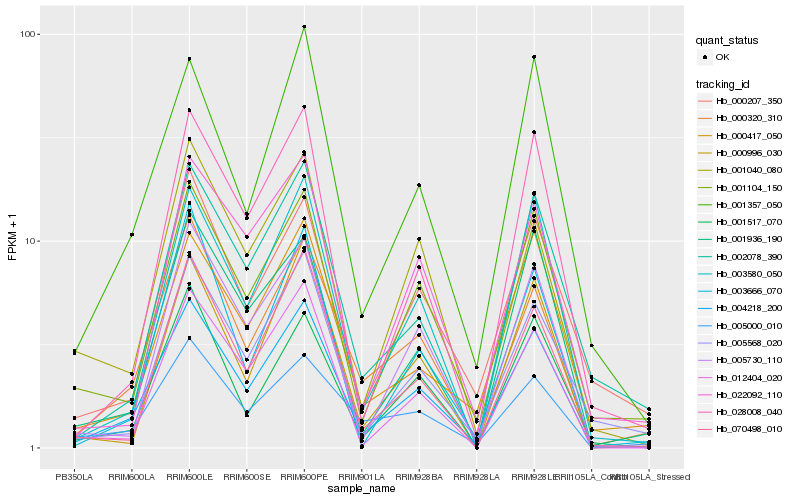
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004218_200 |
0.0 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 2 |
Hb_000320_310 |
0.0997838407 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C [Jatropha curcas] |
| 3 |
Hb_003580_050 |
0.1010286867 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 4 |
Hb_001357_050 |
0.1024332558 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12-like [Jatropha curcas] |
| 5 |
Hb_001104_150 |
0.1069849572 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 6 |
Hb_028008_040 |
0.1146032993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005730_110 |
0.1147193857 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 8 |
Hb_070498_010 |
0.1178383806 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 9 |
Hb_012404_020 |
0.120305178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001517_070 |
0.1213085709 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_005568_020 |
0.12209477 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001936_190 |
0.1230685602 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 13 |
Hb_002078_390 |
0.1281787142 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 14 |
Hb_000996_030 |
0.1293717811 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g63430 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001040_080 |
0.1299366842 |
- |
- |
sphingolipid delta 4 desaturase/C-4 hydroxylase protein des2, putative [Ricinus communis] |
| 16 |
Hb_000417_050 |
0.1309363687 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 17 |
Hb_000207_350 |
0.1310428106 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 18 |
Hb_005000_010 |
0.1314337013 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 19 |
Hb_022092_110 |
0.1329485486 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 20 |
Hb_003666_070 |
0.133007236 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |