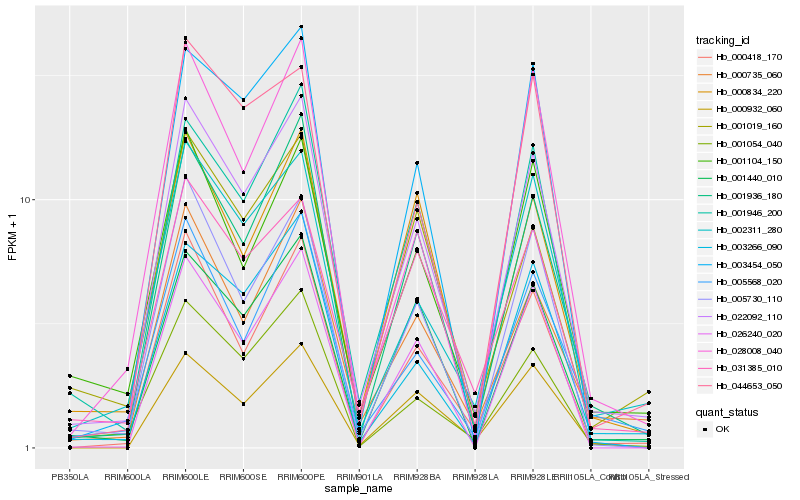
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022092_110 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 2 |
Hb_001936_180 |
0.071812778 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 3 |
Hb_001946_200 |
0.0811555307 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_001440_010 |
0.0886048075 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |
| 5 |
Hb_000735_060 |
0.0894293055 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 6 |
Hb_000418_170 |
0.0895116241 |
- |
- |
PREDICTED: ABC transporter C family member 12-like isoform X4 [Populus euphratica] |
| 7 |
Hb_002311_280 |
0.0899108119 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 8 |
Hb_005730_110 |
0.0919078382 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 9 |
Hb_003266_090 |
0.0932731894 |
- |
- |
PREDICTED: basic 7S globulin 2-like [Jatropha curcas] |
| 10 |
Hb_005568_020 |
0.0981511648 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001054_040 |
0.0985181711 |
- |
- |
hypothetical protein JCGZ_25310 [Jatropha curcas] |
| 12 |
Hb_031385_010 |
0.1034942217 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 13 |
Hb_000834_220 |
0.1043943701 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 14 |
Hb_001104_150 |
0.1063615223 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 15 |
Hb_026240_020 |
0.1067402491 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 16 |
Hb_044653_050 |
0.1112505449 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001019_160 |
0.1120359051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000932_060 |
0.1121123893 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003454_050 |
0.1122752684 |
- |
- |
PREDICTED: probable methyltransferase PMT14 [Jatropha curcas] |
| 20 |
Hb_028008_040 |
0.1124094803 |
- |
- |
conserved hypothetical protein [Ricinus communis] |