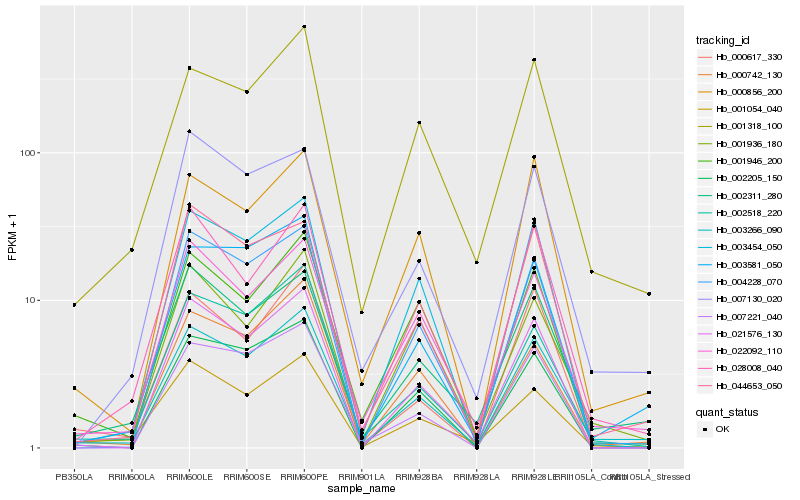
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003266_090 |
0.0 |
- |
- |
PREDICTED: basic 7S globulin 2-like [Jatropha curcas] |
| 2 |
Hb_002311_280 |
0.092448301 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 3 |
Hb_022092_110 |
0.0932731894 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |
| 4 |
Hb_001318_100 |
0.0950968754 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 5 |
Hb_003581_050 |
0.0979451797 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-54 [Jatropha curcas] |
| 6 |
Hb_002518_220 |
0.1004634208 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 7 |
Hb_003454_050 |
0.1015613975 |
- |
- |
PREDICTED: probable methyltransferase PMT14 [Jatropha curcas] |
| 8 |
Hb_028008_040 |
0.1016696559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004228_070 |
0.1019967531 |
- |
- |
PREDICTED: uncharacterized protein LOC103338699 [Prunus mume] |
| 10 |
Hb_021576_130 |
0.1021941198 |
- |
- |
PREDICTED: chorismate mutase 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000742_130 |
0.1043526046 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g30570 [Jatropha curcas] |
| 12 |
Hb_000856_200 |
0.1057059142 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 13 |
Hb_001054_040 |
0.1070807241 |
- |
- |
hypothetical protein JCGZ_25310 [Jatropha curcas] |
| 14 |
Hb_001946_200 |
0.1137649255 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 15 |
Hb_044653_050 |
0.1165895652 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001936_180 |
0.1175597477 |
- |
- |
PREDICTED: protein NSP-INTERACTING KINASE 3 [Jatropha curcas] |
| 17 |
Hb_007130_020 |
0.1175978659 |
- |
- |
- |
| 18 |
Hb_007221_040 |
0.1182663675 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 isoform X2 [Populus euphratica] |
| 19 |
Hb_002205_150 |
0.1198092264 |
- |
- |
RING-H2 finger protein ATL3C, putative [Ricinus communis] |
| 20 |
Hb_000617_330 |
0.1203834563 |
- |
- |
PREDICTED: uncharacterized protein LOC105647507 [Jatropha curcas] |