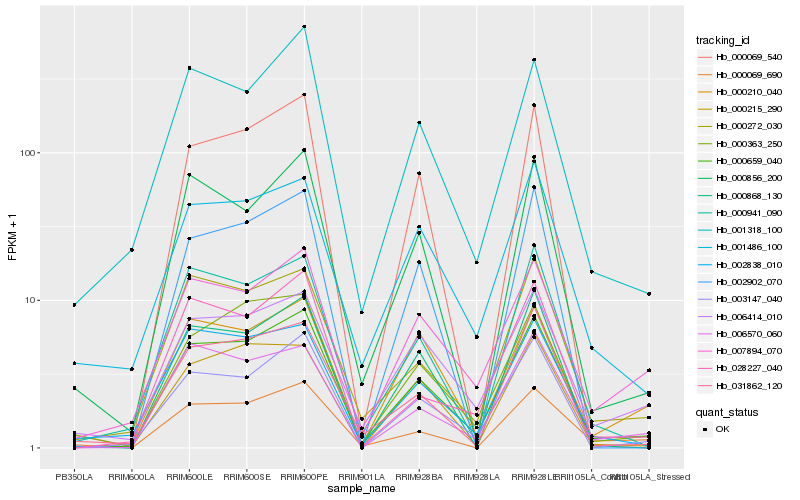
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000659_040 |
0.0 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_003147_040 |
0.0925410364 |
- |
- |
PREDICTED: probable calcium-binding protein CML18 [Jatropha curcas] |
| 3 |
Hb_000363_250 |
0.0997429782 |
- |
- |
PREDICTED: pirin-like protein [Malus domestica] |
| 4 |
Hb_000272_030 |
0.1068382805 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001318_100 |
0.1068397632 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 6 |
Hb_000856_200 |
0.1087664157 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 7 |
Hb_007894_070 |
0.1094705184 |
- |
- |
PREDICTED: uncharacterized protein LOC105632955 [Jatropha curcas] |
| 8 |
Hb_000941_090 |
0.1103055082 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 9 |
Hb_002838_010 |
0.1120549625 |
- |
- |
AGPase large subunit protein [Hevea brasiliensis] |
| 10 |
Hb_006414_010 |
0.1120607259 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 11 |
Hb_031862_120 |
0.1126554734 |
- |
- |
PREDICTED: root phototropism protein 3 [Jatropha curcas] |
| 12 |
Hb_000069_540 |
0.1165070554 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Jatropha curcas] |
| 13 |
Hb_000868_130 |
0.118025211 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 14 |
Hb_006570_060 |
0.1181804393 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 15 |
Hb_028227_040 |
0.1184422963 |
- |
- |
PREDICTED: uncharacterized protein LOC105645005 [Jatropha curcas] |
| 16 |
Hb_000215_290 |
0.1210051173 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001486_100 |
0.121627681 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 18 |
Hb_000210_040 |
0.1226060037 |
- |
- |
PREDICTED: putative HVA22-like protein g [Jatropha curcas] |
| 19 |
Hb_000069_690 |
0.1227823564 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF6 [Jatropha curcas] |
| 20 |
Hb_002902_070 |
0.1242887157 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |