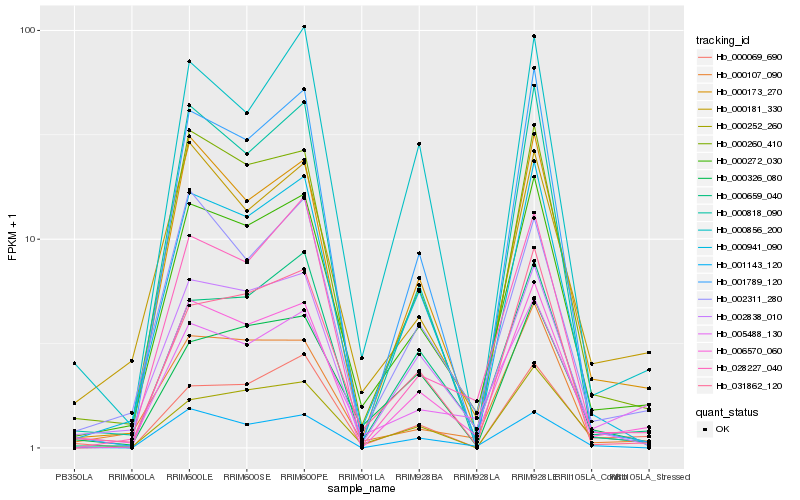
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000272_030 |
0.0 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 2 |
Hb_006570_060 |
0.0881940508 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 3 |
Hb_000260_410 |
0.0904489491 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 4 |
Hb_000941_090 |
0.092588041 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 5 |
Hb_002838_010 |
0.0964386772 |
- |
- |
AGPase large subunit protein [Hevea brasiliensis] |
| 6 |
Hb_000173_270 |
0.0998854651 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000107_090 |
0.1034978134 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 8 |
Hb_031862_120 |
0.1040793219 |
- |
- |
PREDICTED: root phototropism protein 3 [Jatropha curcas] |
| 9 |
Hb_000659_040 |
0.1068382805 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000181_330 |
0.1130151456 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 11 |
Hb_028227_040 |
0.1180426483 |
- |
- |
PREDICTED: uncharacterized protein LOC105645005 [Jatropha curcas] |
| 12 |
Hb_002311_280 |
0.1189387416 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 13 |
Hb_005488_130 |
0.122494908 |
- |
- |
transporter, putative [Ricinus communis] |
| 14 |
Hb_000252_260 |
0.1234258469 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 15 |
Hb_000326_080 |
0.1265579372 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 16 |
Hb_000856_200 |
0.1303672295 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 17 |
Hb_000069_690 |
0.1319837035 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF6 [Jatropha curcas] |
| 18 |
Hb_001143_120 |
0.1320742068 |
- |
- |
PREDICTED: uncharacterized protein LOC105638955 [Jatropha curcas] |
| 19 |
Hb_000818_090 |
0.1331990035 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 20 |
Hb_001789_120 |
0.1333039691 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |