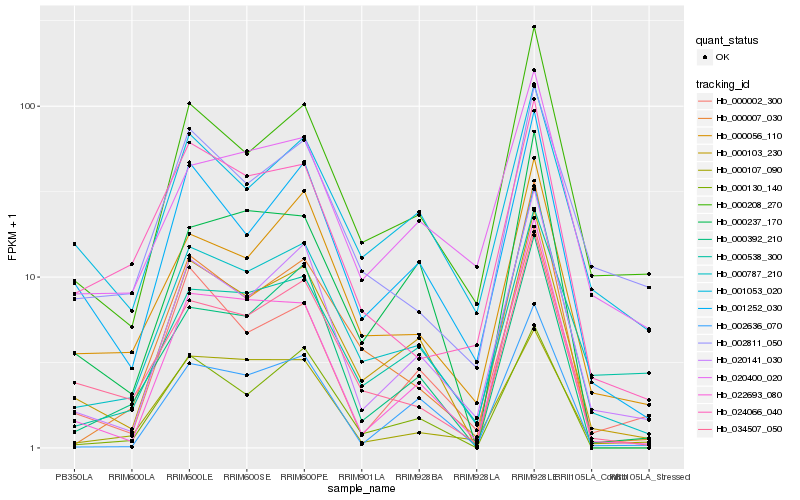
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000787_210 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000103_230 |
0.081131258 |
- |
- |
PREDICTED: chlorophyllase-2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_020141_030 |
0.0933143133 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 4 |
Hb_000056_110 |
0.100778719 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |
| 5 |
Hb_000208_270 |
0.1033964992 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 6 |
Hb_034507_050 |
0.1082069817 |
- |
- |
PREDICTED: uncharacterized protein LOC105646633 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001252_030 |
0.1156803564 |
- |
- |
Serine/threonine-protein kinase Nek8, putative [Ricinus communis] |
| 8 |
Hb_000007_030 |
0.1197758144 |
- |
- |
PREDICTED: chaperonin 60 subunit beta 4, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000130_140 |
0.1290622426 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639203 isoform X1 [Jatropha curcas] |
| 10 |
Hb_022693_080 |
0.1301878952 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 11 |
Hb_020400_020 |
0.1338542991 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 12 |
Hb_000392_210 |
0.1341798436 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0004s04600g [Populus trichocarpa] |
| 13 |
Hb_000107_090 |
0.1380484944 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 14 |
Hb_000538_300 |
0.1388906642 |
- |
- |
PREDICTED: WAT1-related protein At3g02690, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001053_020 |
0.1395196026 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 16 |
Hb_024066_040 |
0.1397194809 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 17 |
Hb_000237_170 |
0.1400957203 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 6 [Jatropha curcas] |
| 18 |
Hb_002811_050 |
0.1409828404 |
- |
- |
PREDICTED: uncharacterized protein LOC105647652 [Jatropha curcas] |
| 19 |
Hb_000002_300 |
0.1418456882 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 20 |
Hb_002636_070 |
0.1419036107 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |