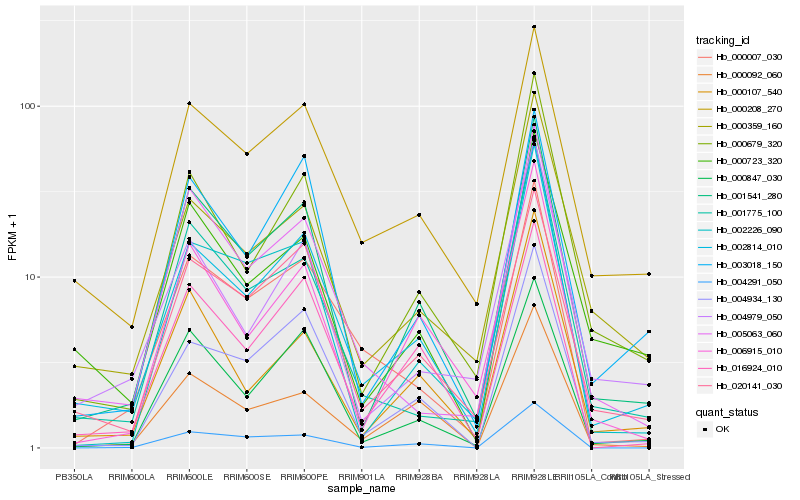
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002814_010 |
0.0 |
- |
- |
PREDICTED: adenylate kinase 5, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001541_280 |
0.0935809608 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 3 |
Hb_004934_130 |
0.094966254 |
- |
- |
12-oxophytodienoate reductase 2 isoform 1 [Theobroma cacao] |
| 4 |
Hb_000679_320 |
0.0992894973 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105136146 [Populus euphratica] |
| 5 |
Hb_020141_030 |
0.1081370658 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 6 |
Hb_000208_270 |
0.1103795909 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 7 |
Hb_000359_160 |
0.1125061437 |
- |
- |
PREDICTED: protein CAJ1 [Jatropha curcas] |
| 8 |
Hb_000847_030 |
0.1211905243 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 9 |
Hb_002226_090 |
0.1240921075 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 10 |
Hb_001775_100 |
0.1247439441 |
- |
- |
PREDICTED: probable 2-carboxy-D-arabinitol-1-phosphatase [Jatropha curcas] |
| 11 |
Hb_016924_010 |
0.1251116201 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000007_030 |
0.1255281077 |
- |
- |
PREDICTED: chaperonin 60 subunit beta 4, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_004979_050 |
0.1256914757 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 14 |
Hb_003018_150 |
0.1293811511 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 15 |
Hb_006915_010 |
0.1304909056 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 16 |
Hb_000107_540 |
0.1307091086 |
- |
- |
PREDICTED: uncharacterized protein LOC105635335 [Jatropha curcas] |
| 17 |
Hb_000092_060 |
0.1323223112 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 18 |
Hb_005063_060 |
0.1327881723 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105647704 [Jatropha curcas] |
| 19 |
Hb_004291_050 |
0.132955478 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 20 |
Hb_000723_320 |
0.1331155266 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |