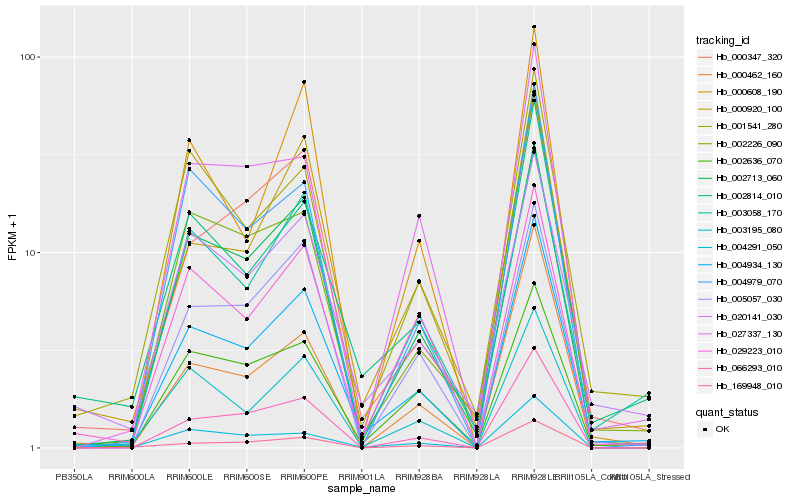
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004934_130 |
0.0 |
- |
- |
12-oxophytodienoate reductase 2 isoform 1 [Theobroma cacao] |
| 2 |
Hb_002814_010 |
0.094966254 |
- |
- |
PREDICTED: adenylate kinase 5, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002713_060 |
0.0977099597 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 4 |
Hb_000920_100 |
0.1050700527 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_066293_010 |
0.1068318831 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 6 |
Hb_002226_090 |
0.110612492 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 7 |
Hb_001541_280 |
0.1145815694 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 8 |
Hb_000462_160 |
0.1158303233 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_002636_070 |
0.1184660294 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 10 |
Hb_003058_170 |
0.1191130487 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 11 |
Hb_029223_010 |
0.1193425068 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 12 |
Hb_020141_030 |
0.1196170388 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 13 |
Hb_005057_030 |
0.1197008594 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 14 |
Hb_003195_080 |
0.119760353 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 15 |
Hb_004291_050 |
0.120095501 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 16 |
Hb_000347_320 |
0.1201451401 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 17 |
Hb_027337_130 |
0.1205872747 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 18 |
Hb_169948_010 |
0.1215652421 |
- |
- |
hypothetical protein VITISV_006524 [Vitis vinifera] |
| 19 |
Hb_000608_190 |
0.1225653687 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 20 |
Hb_004979_070 |
0.1232235944 |
- |
- |
wall-associated receptor kinase galacturonan-binding protein [Medicago truncatula] |