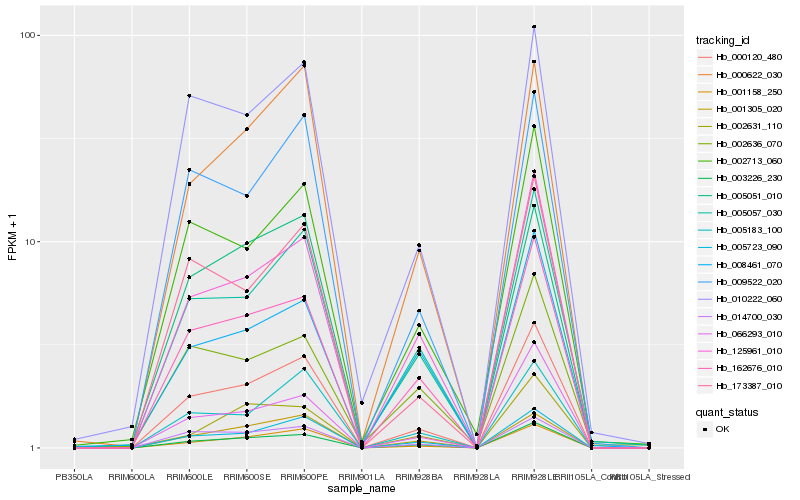
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003226_230 |
0.0 |
- |
- |
PREDICTED: transposon Ty3-G Gag-Pol polyprotein [Phoenix dactylifera] |
| 2 |
Hb_162676_010 |
0.0338066371 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_125961_010 |
0.0453146902 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_000120_480 |
0.0486449189 |
- |
- |
- |
| 5 |
Hb_005057_030 |
0.0839057823 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 6 |
Hb_005723_090 |
0.0861829085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_066293_010 |
0.0877413985 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 8 |
Hb_008461_070 |
0.0986307972 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001158_250 |
0.1015889744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002713_060 |
0.1049700272 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 11 |
Hb_014700_030 |
0.105071245 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 12 |
Hb_002631_110 |
0.1112105408 |
- |
- |
PREDICTED: cytochrome P450 71D9-like [Fragaria vesca subsp. vesca] |
| 13 |
Hb_001305_020 |
0.1164185833 |
- |
- |
PREDICTED: uncharacterized protein LOC102623247 [Citrus sinensis] |
| 14 |
Hb_010222_060 |
0.1181641758 |
- |
- |
PREDICTED: acyltransferase-like protein At1g54570, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_000622_030 |
0.1207211532 |
- |
- |
PREDICTED: hydroquinone glucosyltransferase [Jatropha curcas] |
| 16 |
Hb_005051_010 |
0.1214119037 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_002636_070 |
0.1230189811 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 18 |
Hb_005183_100 |
0.1240785465 |
- |
- |
hypothetical protein CICLE_v10002898mg [Citrus clementina] |
| 19 |
Hb_009522_020 |
0.1240868826 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |
| 20 |
Hb_173387_010 |
0.1243189359 |
- |
- |
hypothetical protein POPTR_0017s13190g, partial [Populus trichocarpa] |